Histogram of correlated \(z\) scores, Part 1
Lei Sun
2017-03-06
Last updated: 2017-12-21
Code version: 6e42447
Introduction
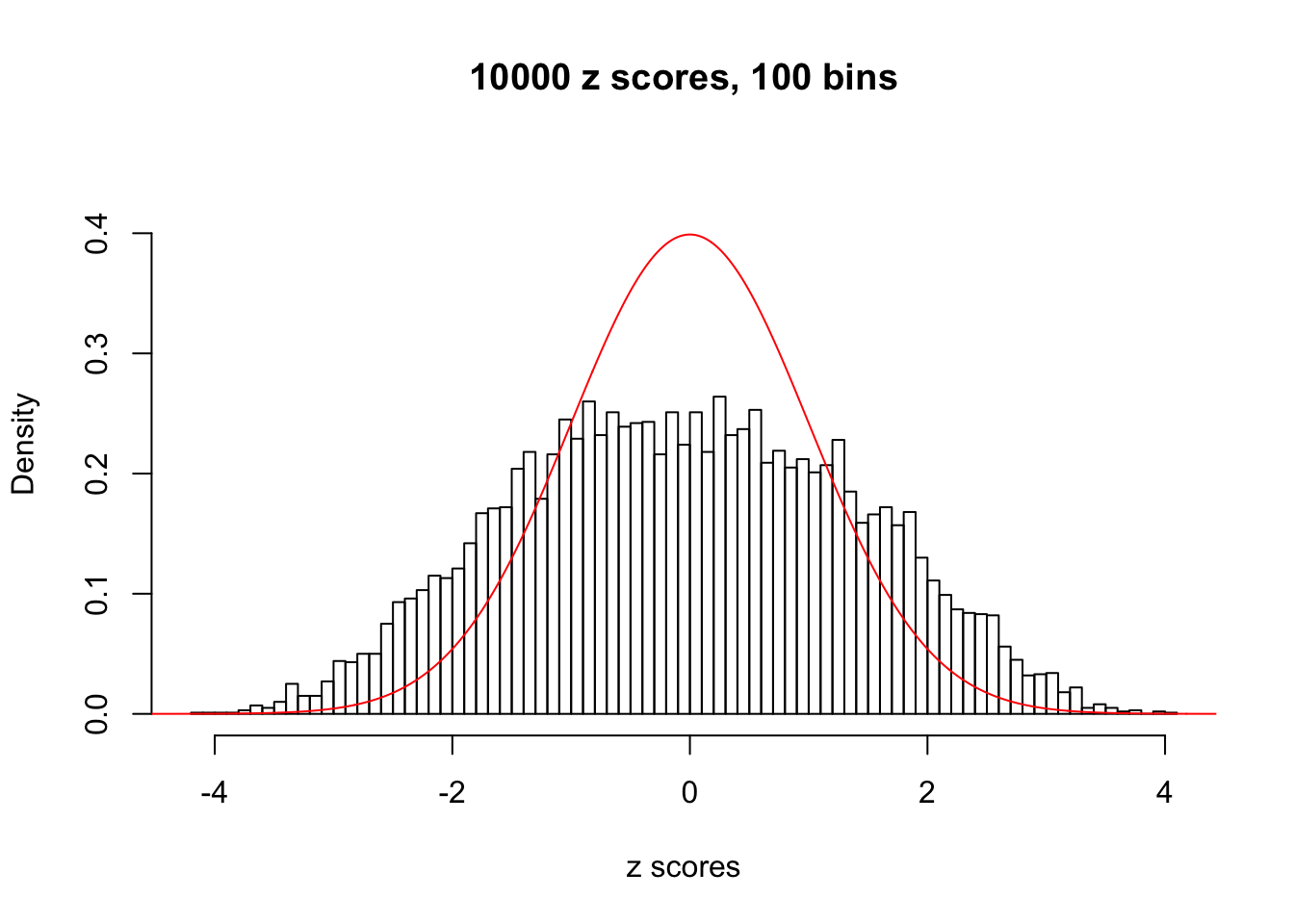
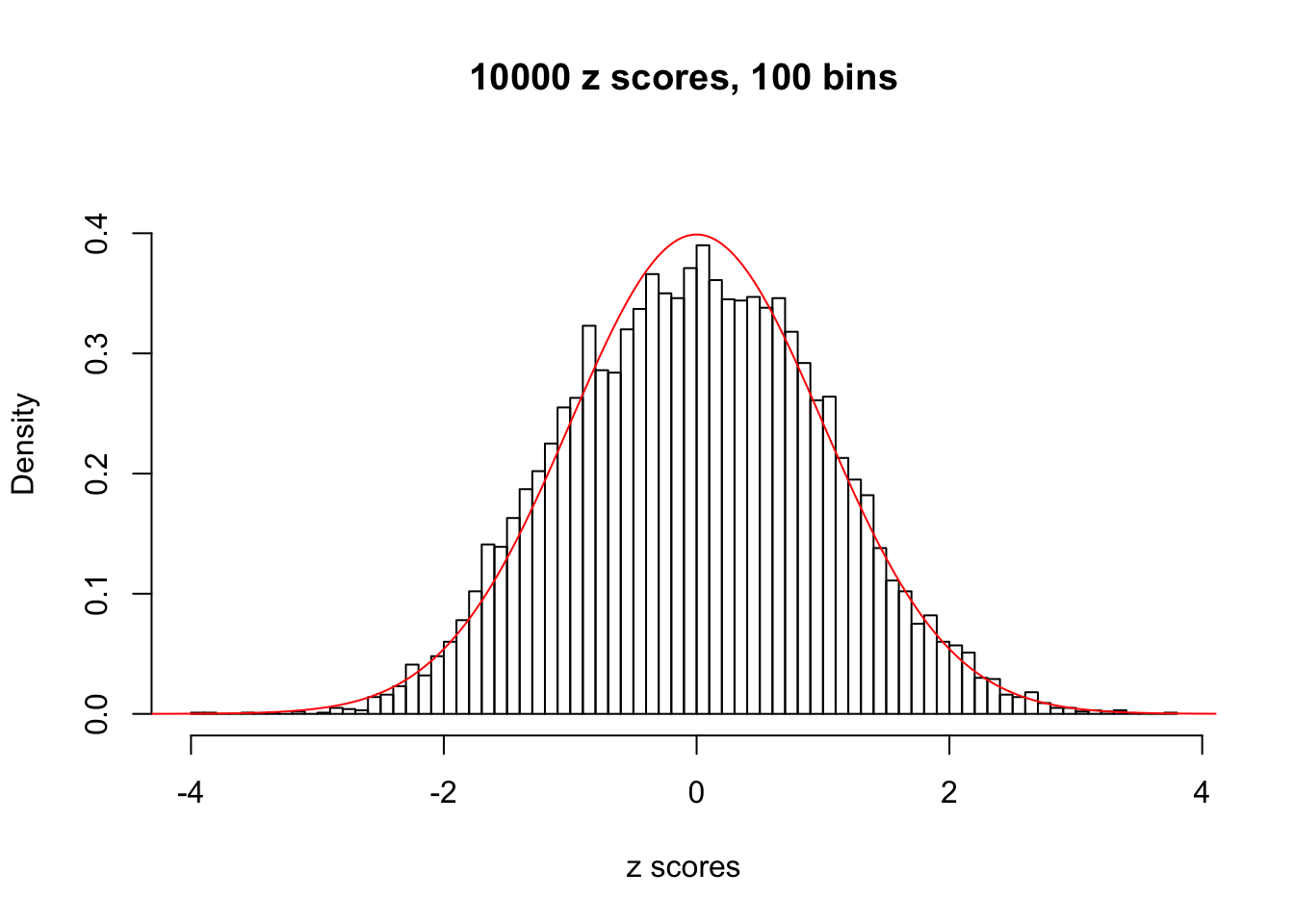
Efron 2010 and Schwartzman’s comment brings to the center the question “what’s the behavior of \(z\) scores under correlation?” Schwartzman pointed out in theory that “the observed histogram is more likely to be narrow than wide, and that it cannot be too wide before it becomes bimodal.” Let’s take a look if this result holds true under our simulation scheme with GTex/Liver data.
Histograms on randomly sampled data sets
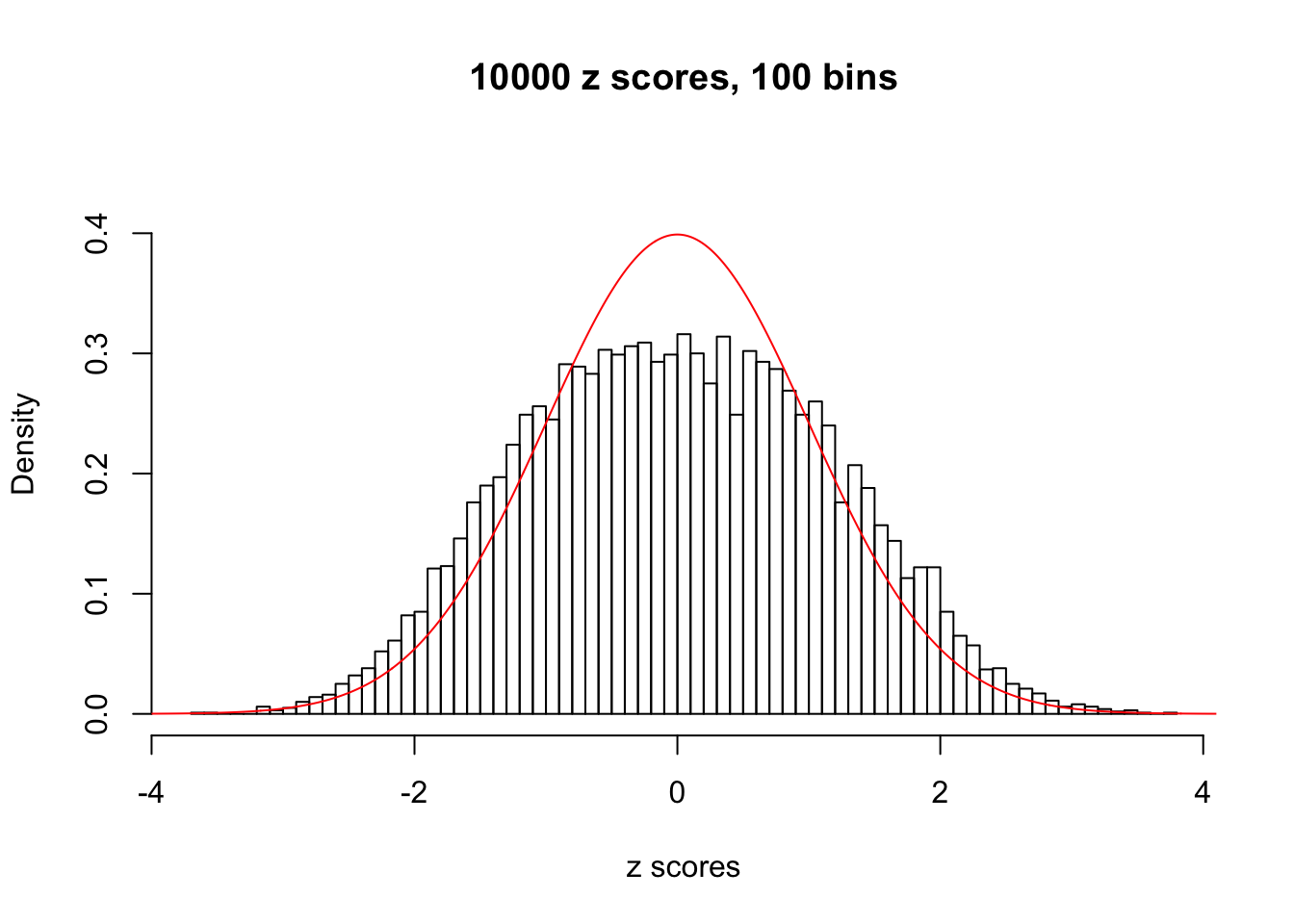
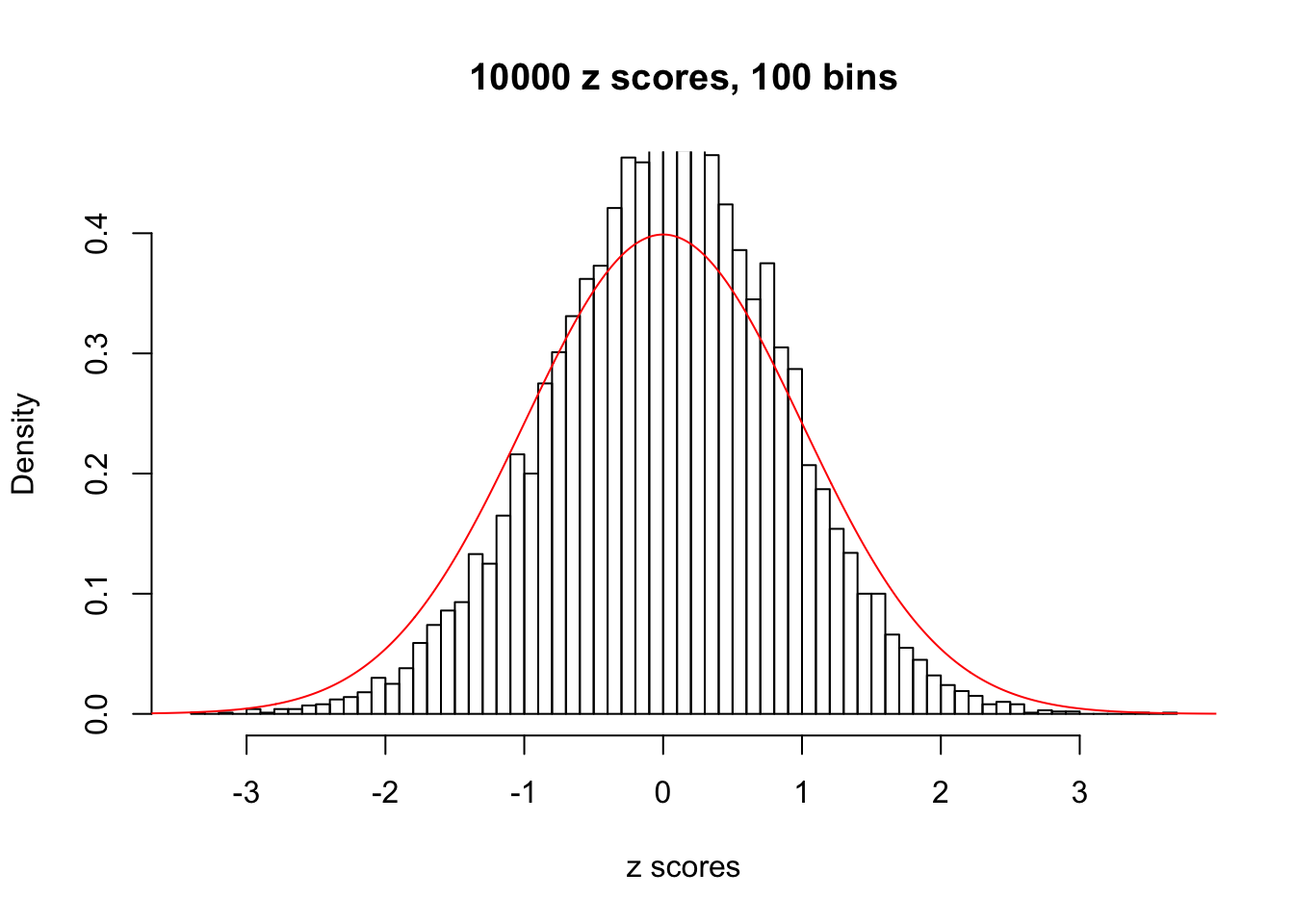
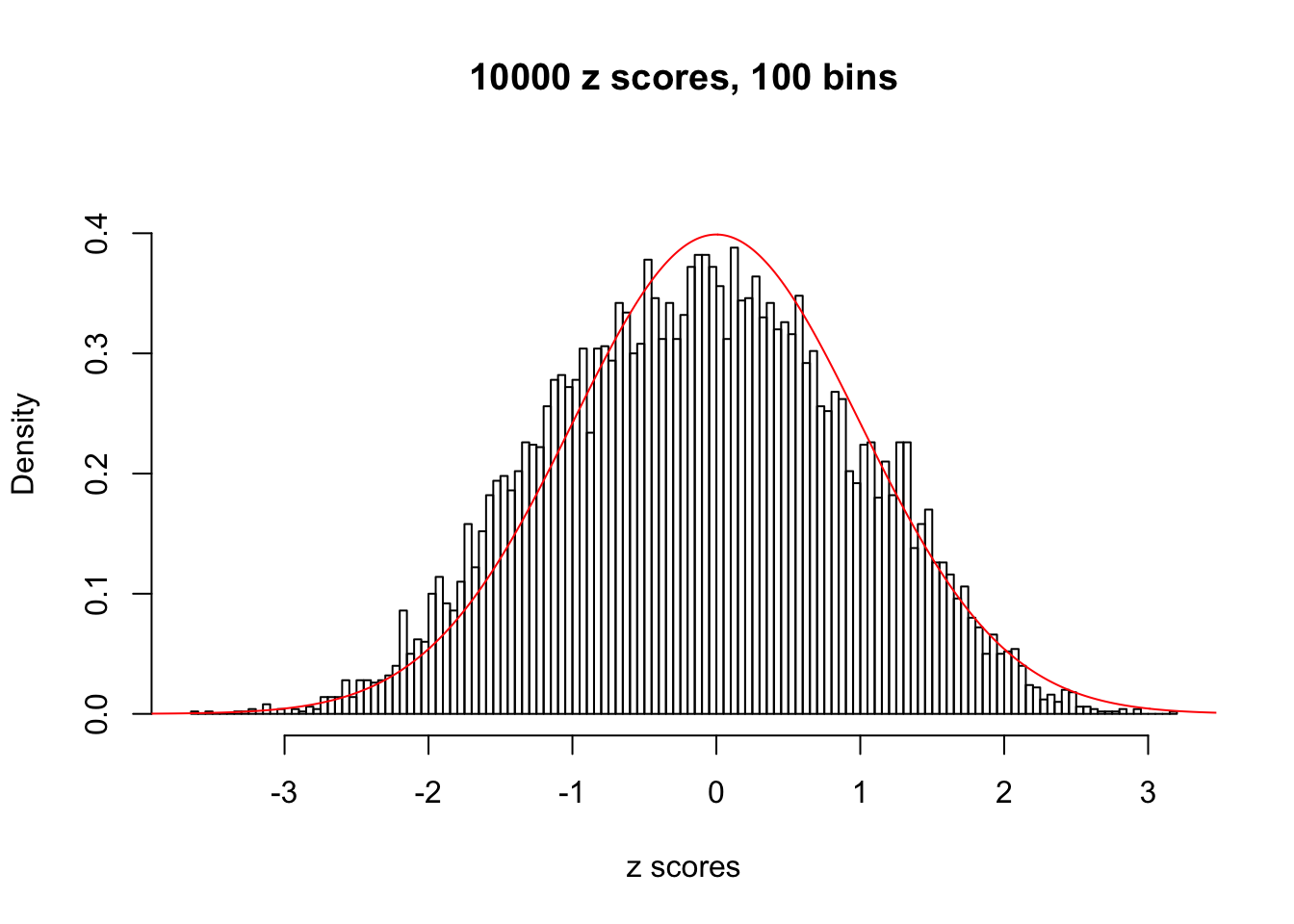
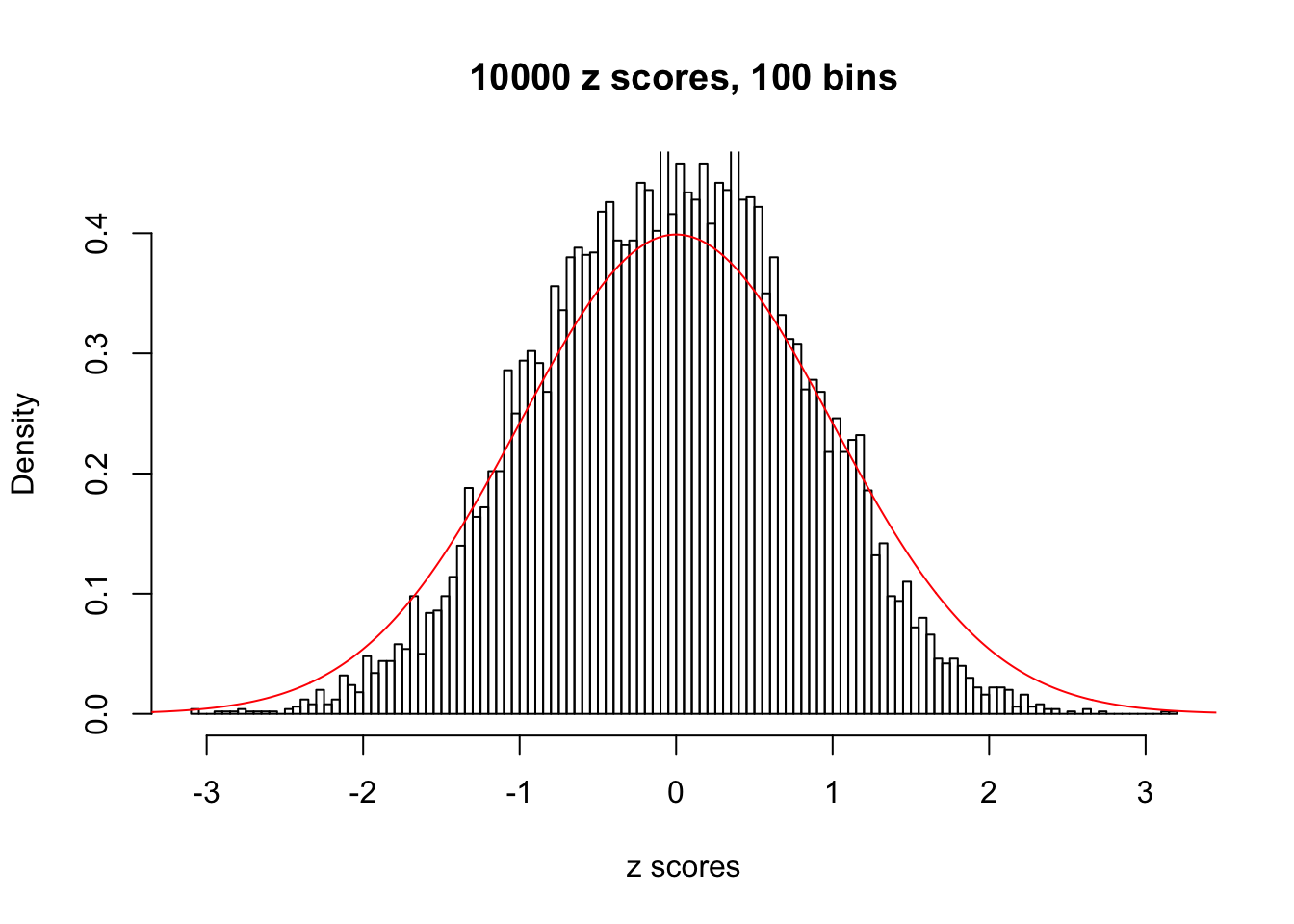
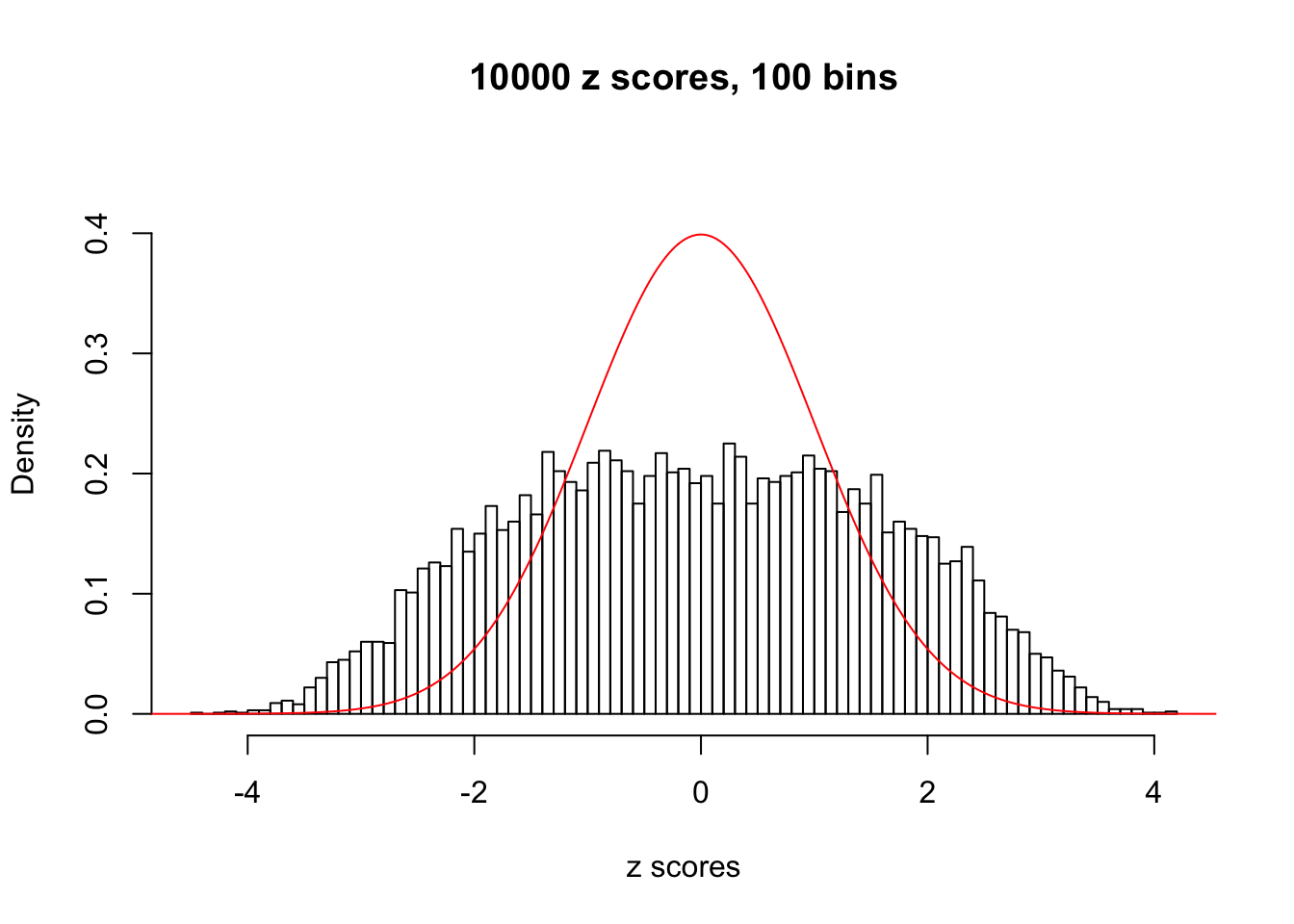
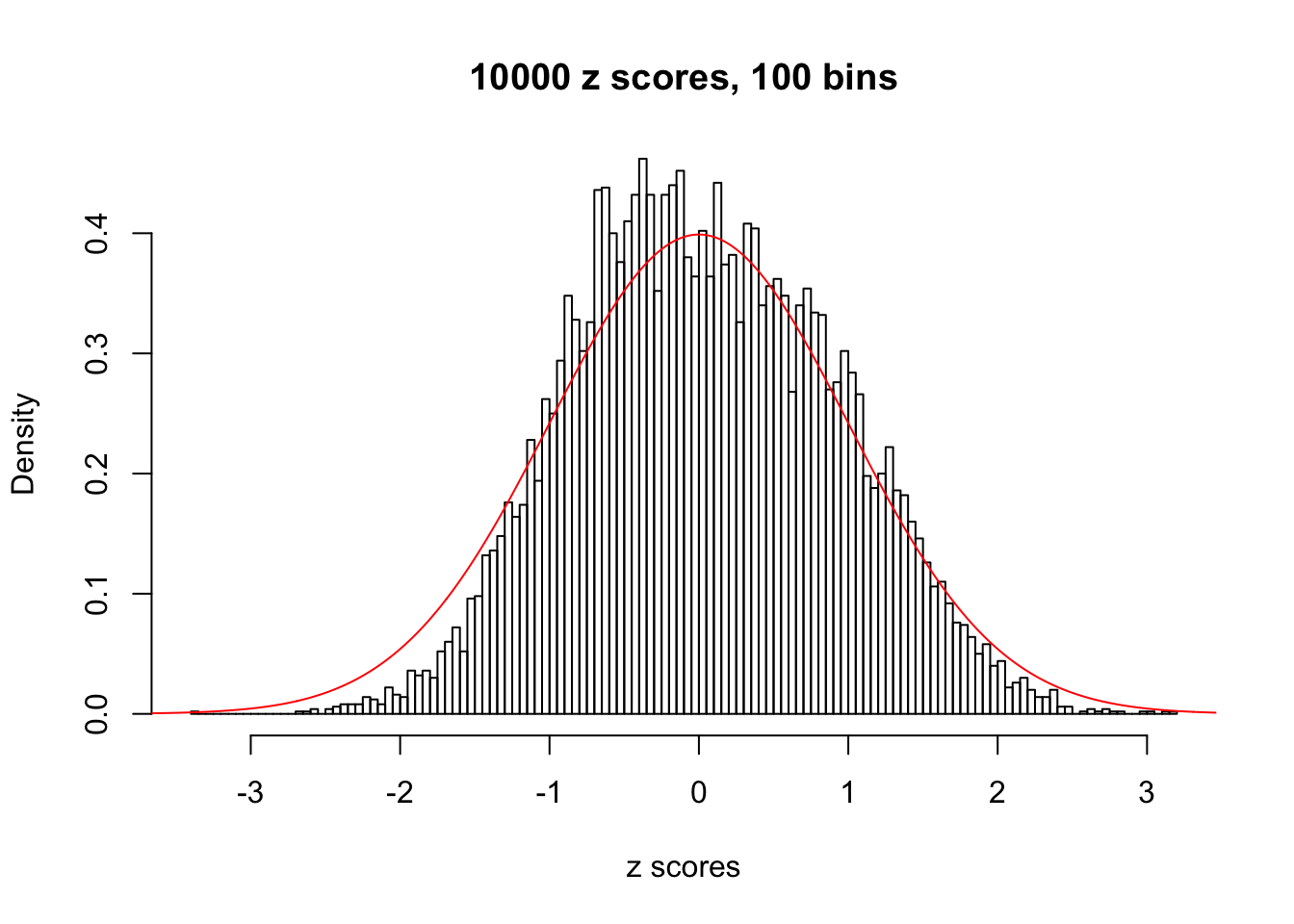
z = read.table("../output/z_null_liver_777.txt")We randomly selected 20 data sets, each with \(10000\) z scores, generated by the null simulation pipeline. Based on each data set we plot two histograms, one using default number of bins and the other \(100\) bins. The red line indicates the density of \(N(0, 1)\).
set.seed(777)
sample_z = sort(sample(dim(z)[1], 20))
x = seq(- 10, 10, 0.01)
y = dnorm(x)
for (i in sample_z) {
cat("Data Set", i)
hist(as.numeric(z[i, ]), xlab = "z scores", freq = FALSE, ylim = c(0, 0.45), main = "10000 z scores, default")
lines(x, y, col = "red")
hist(as.numeric(z[i, ]), xlab = "z scores", freq = FALSE, ylim = c(0, 0.45), nclass = 100, main = "10000 z scores, 100 bins")
lines(x, y, col = "red")
}Data Set 11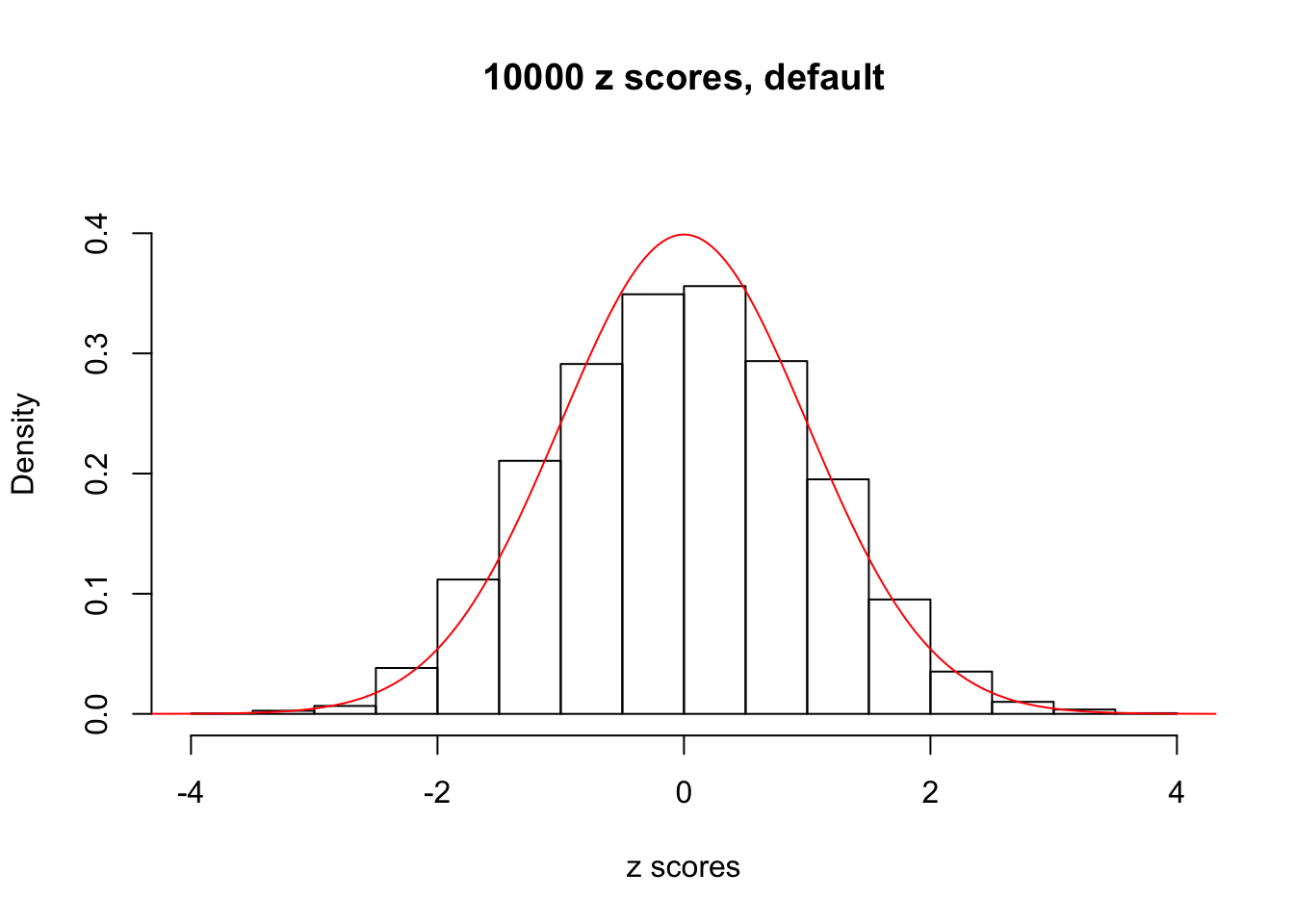
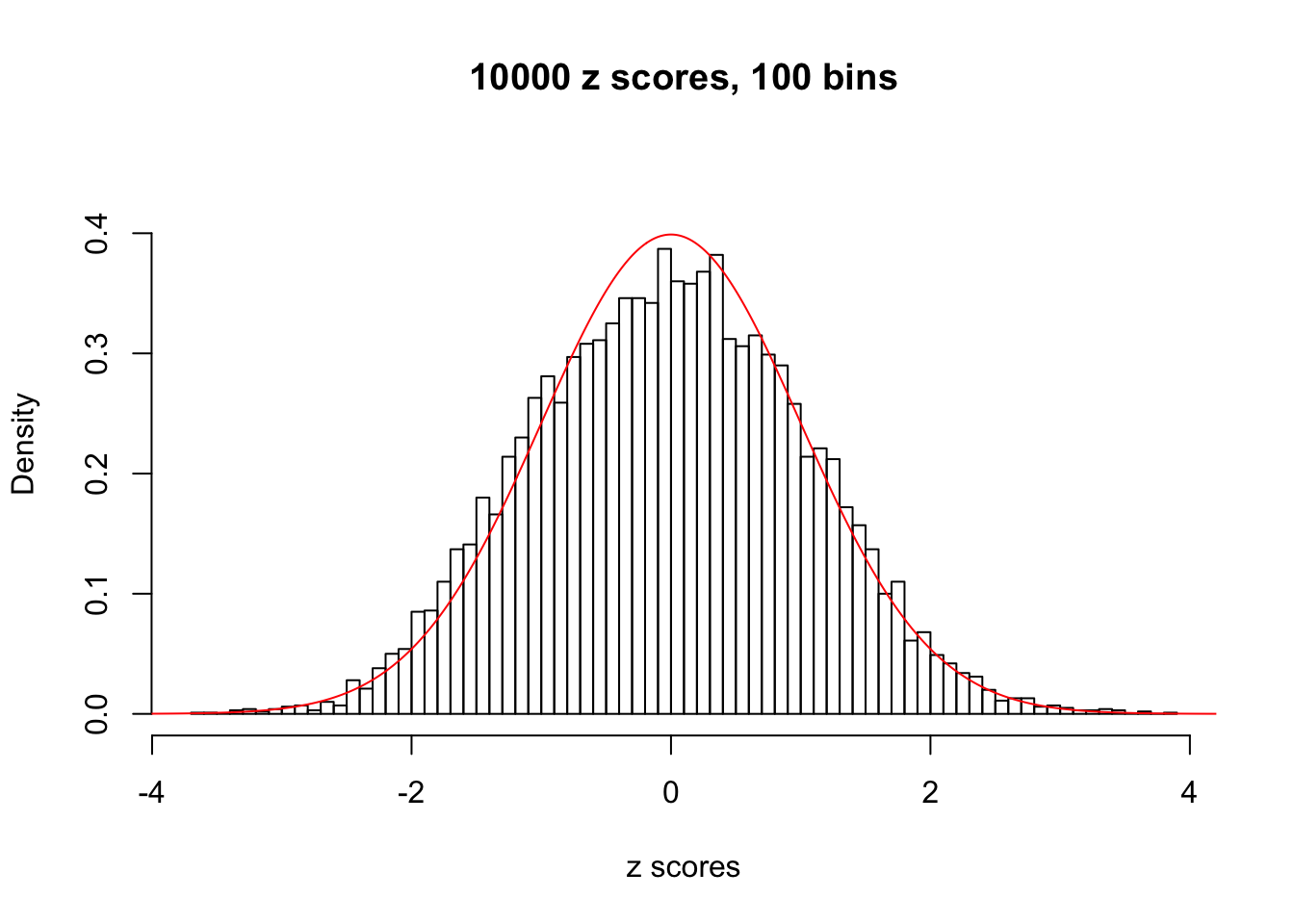
Data Set 103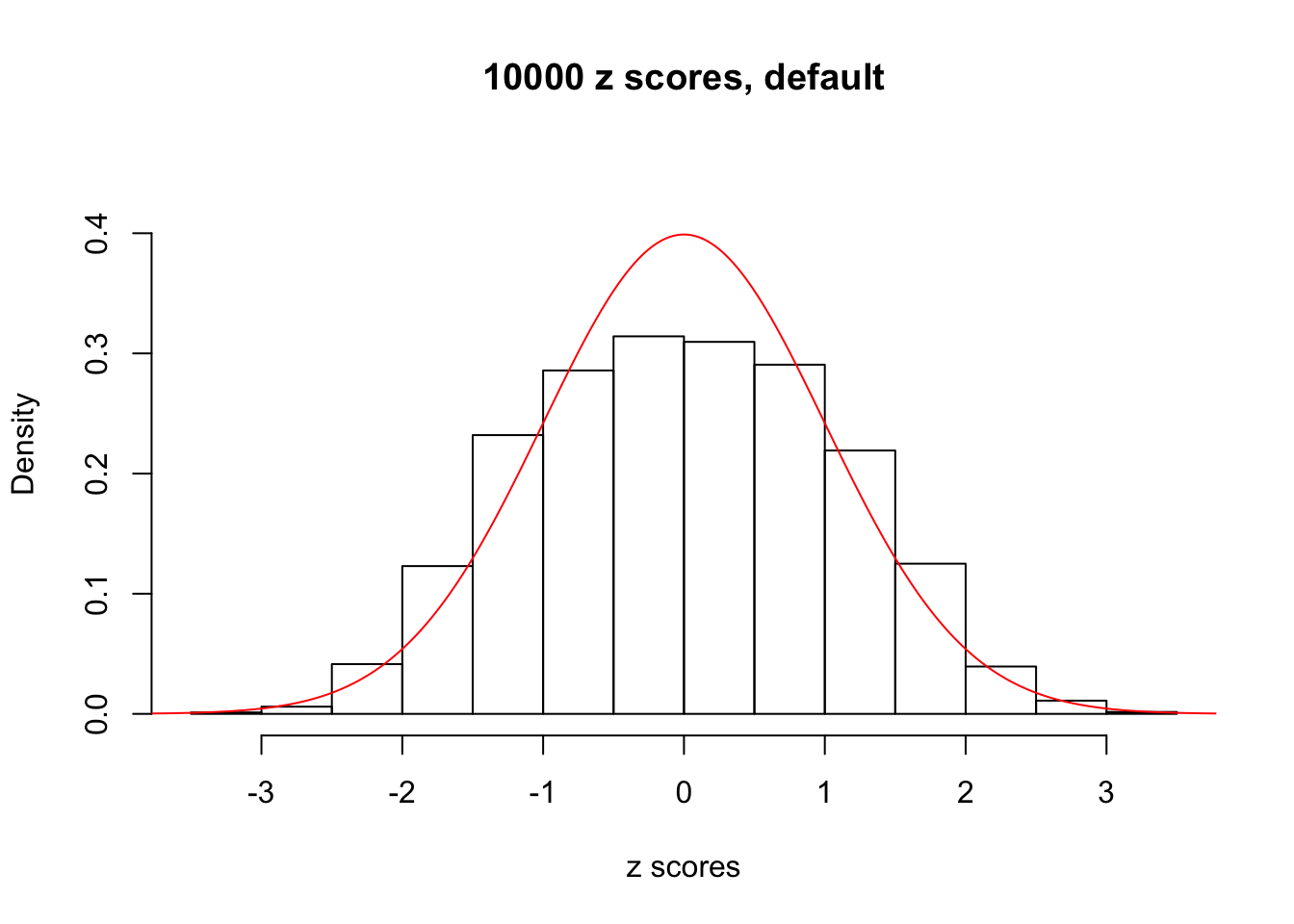
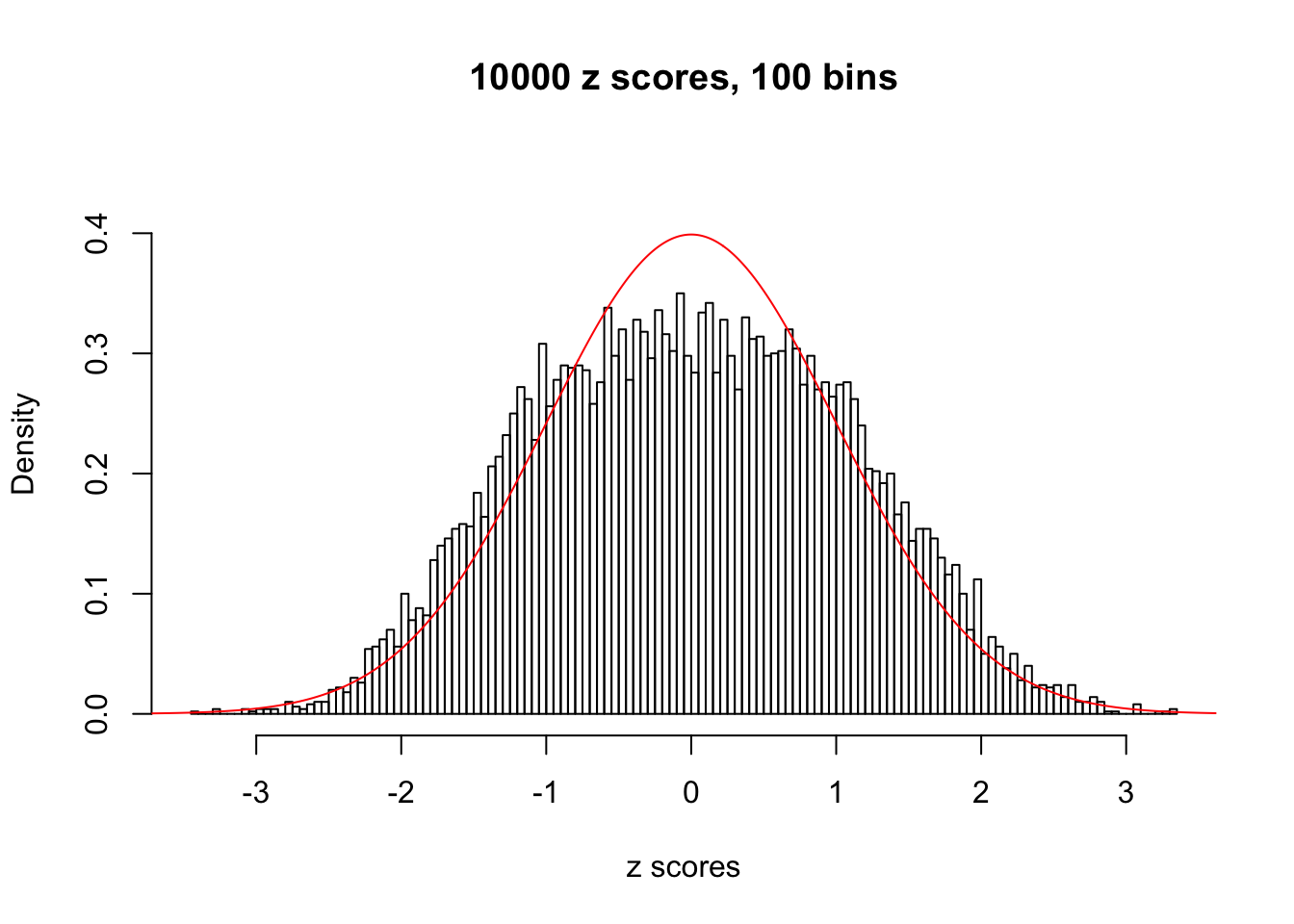
Data Set 171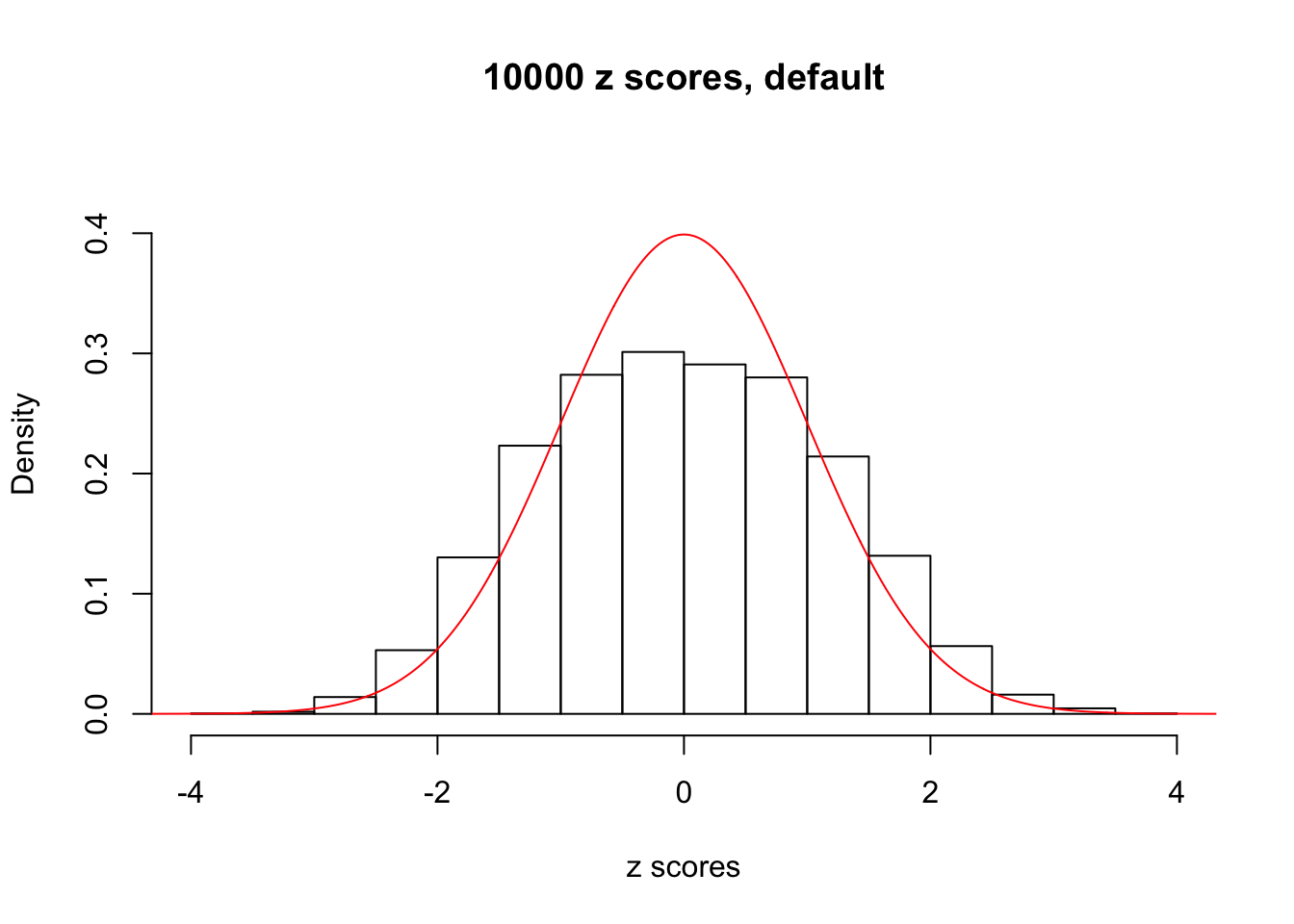
Data Set 247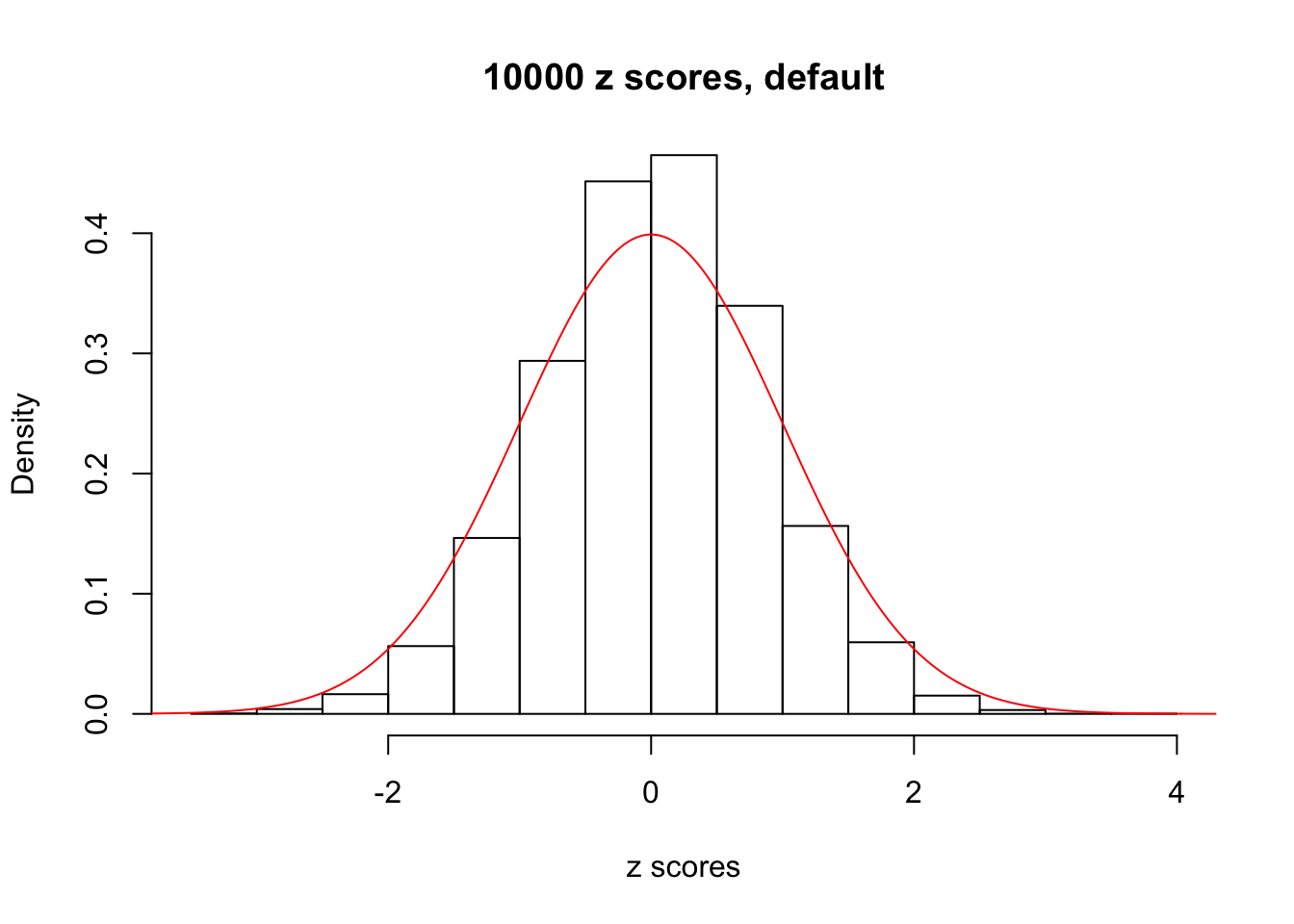
Data Set 343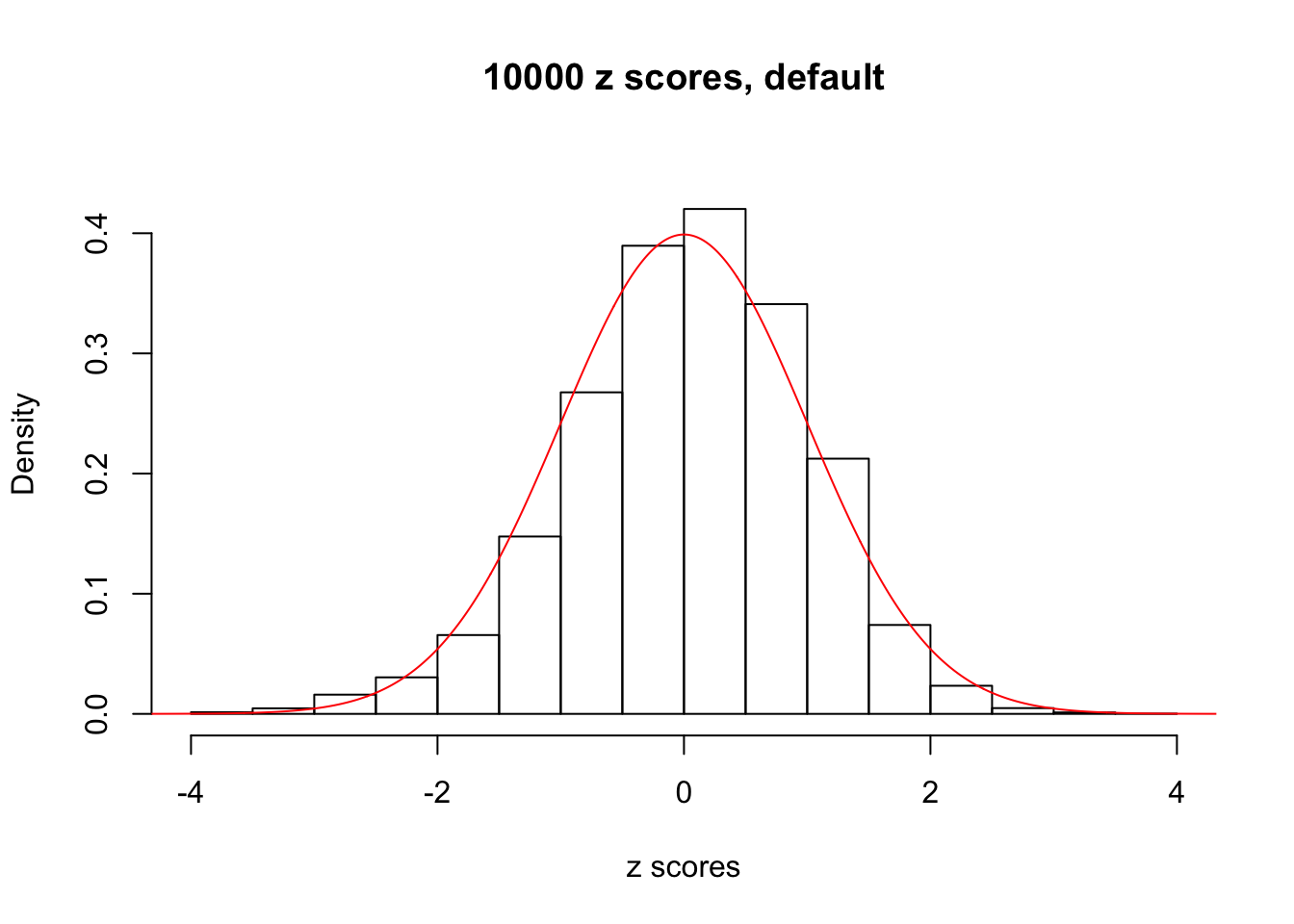
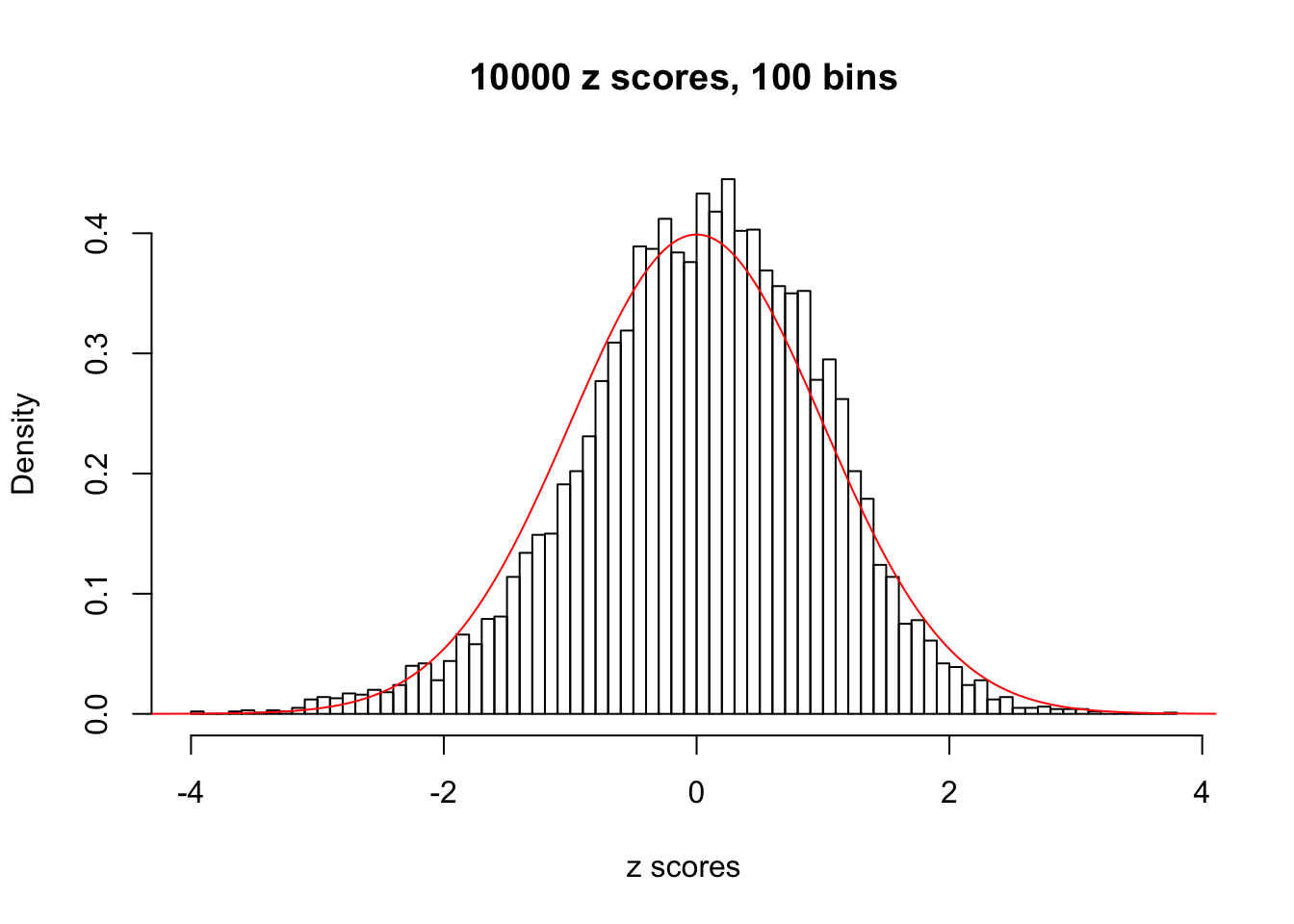
Data Set 345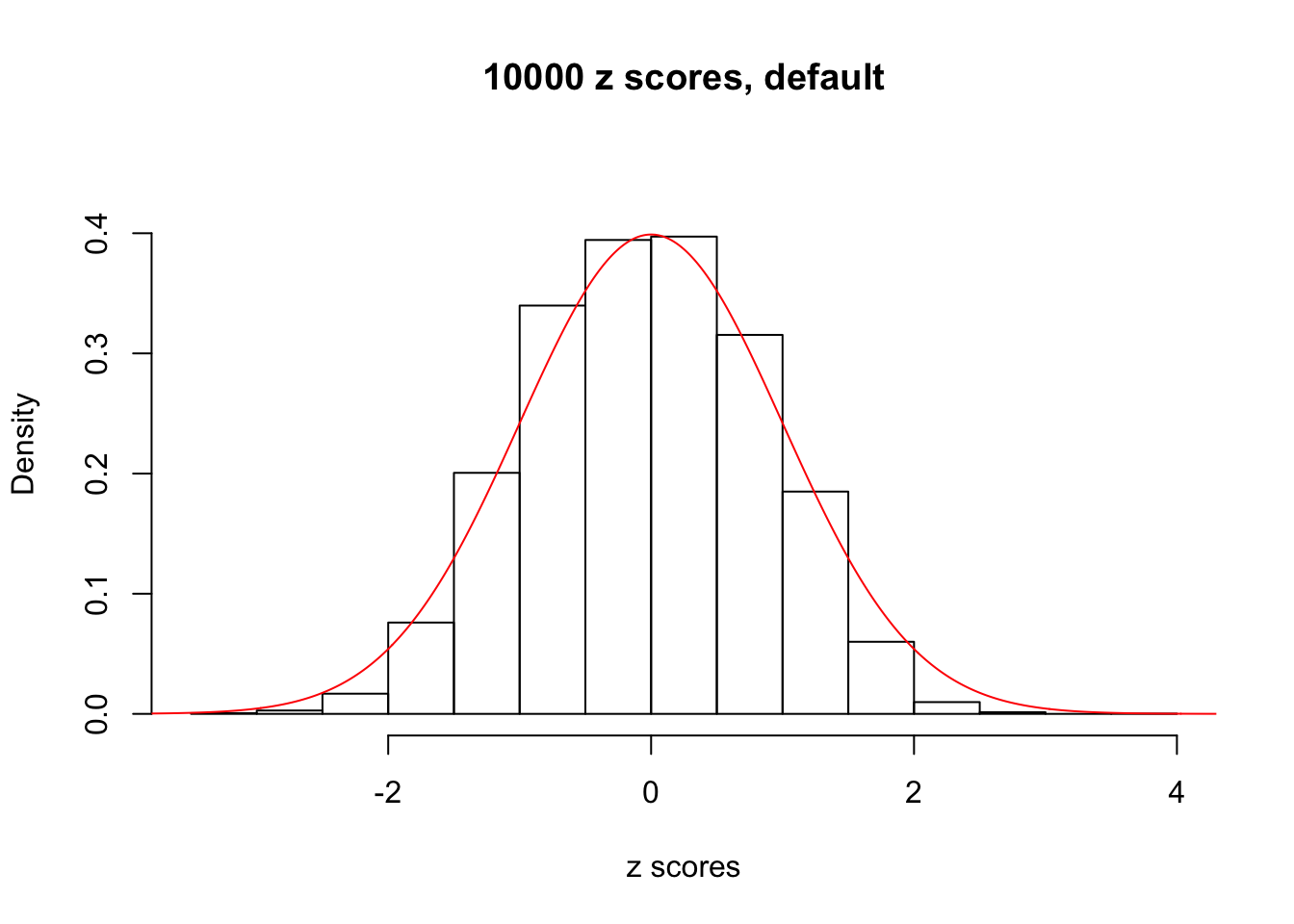
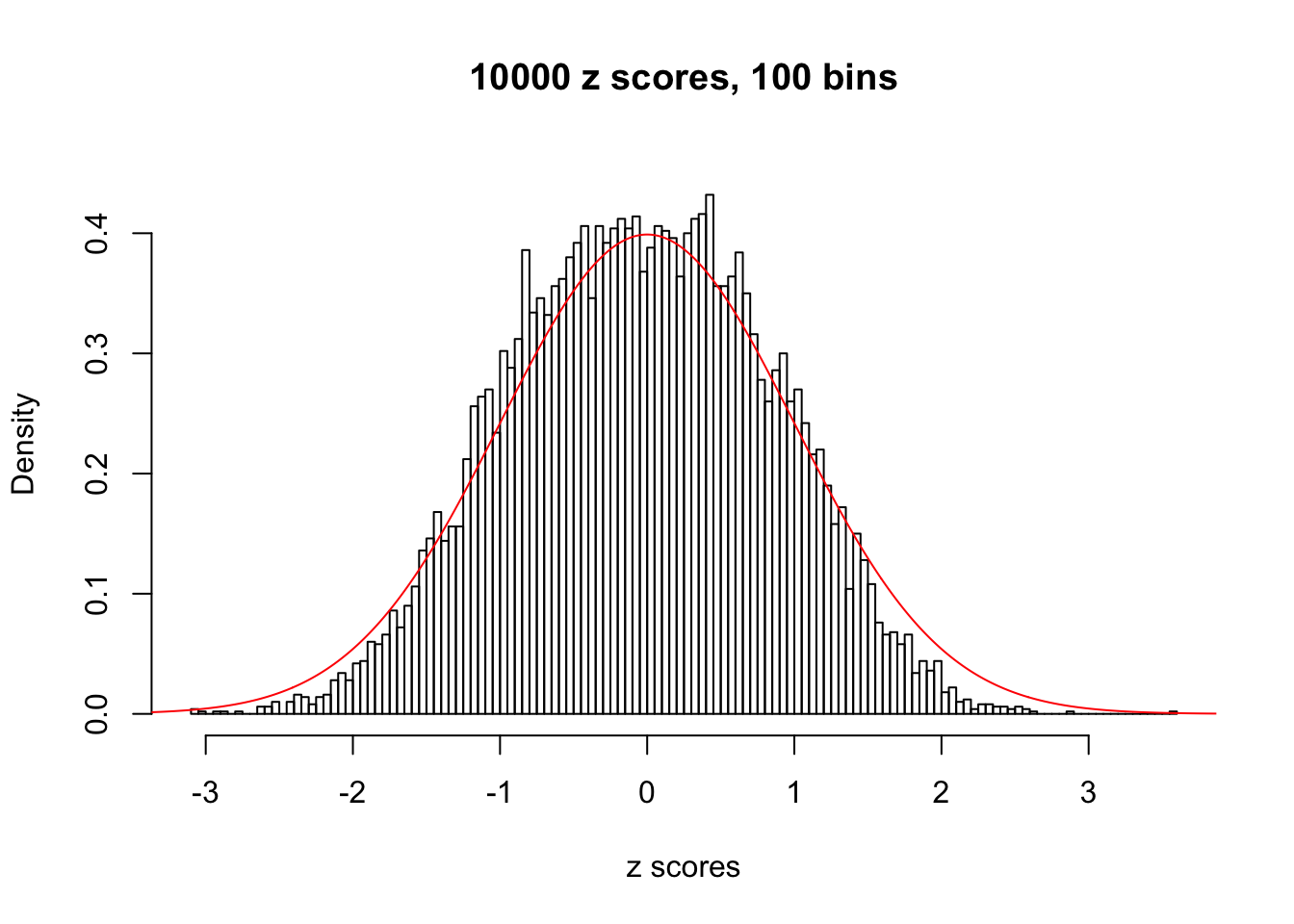
Data Set 347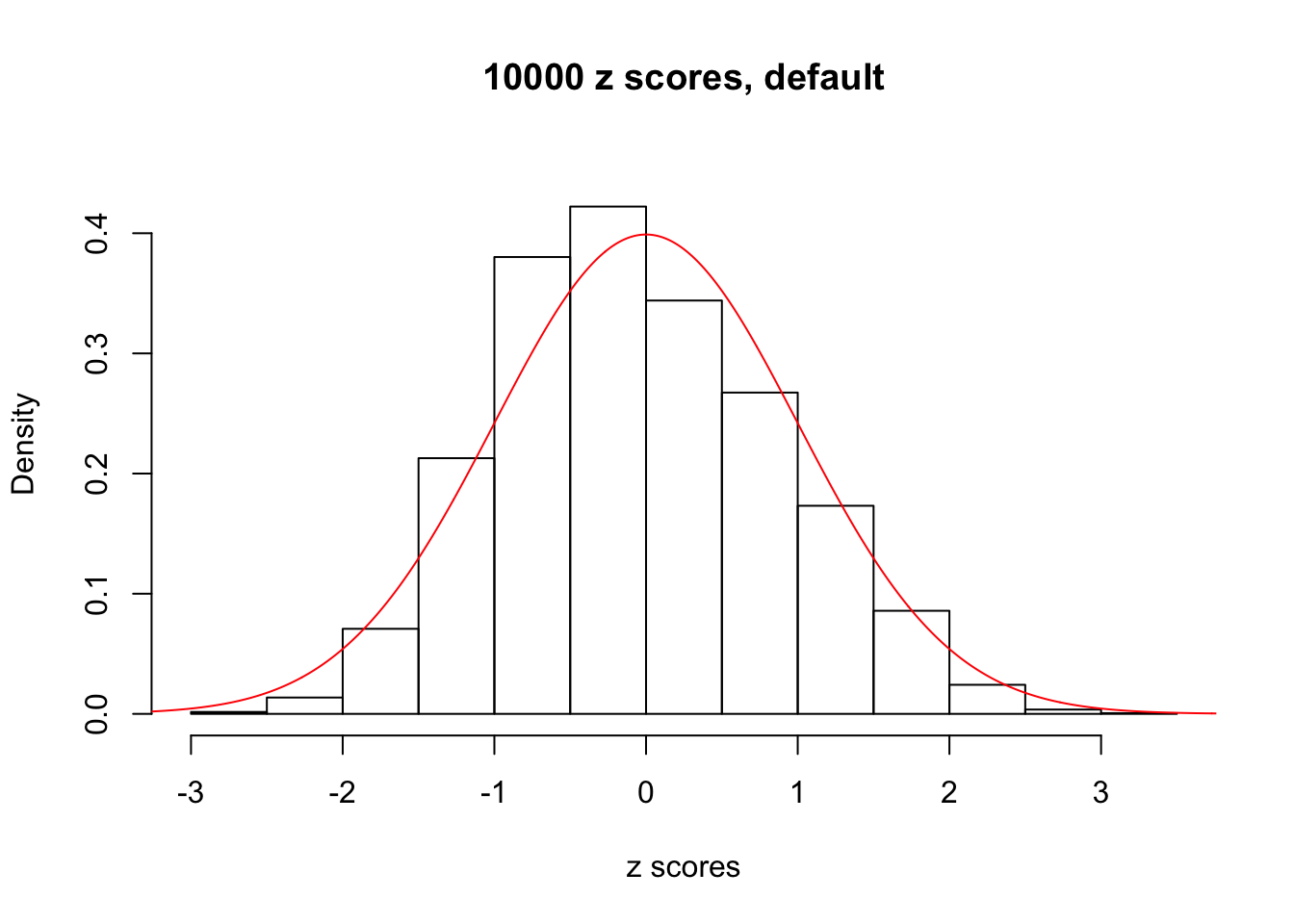
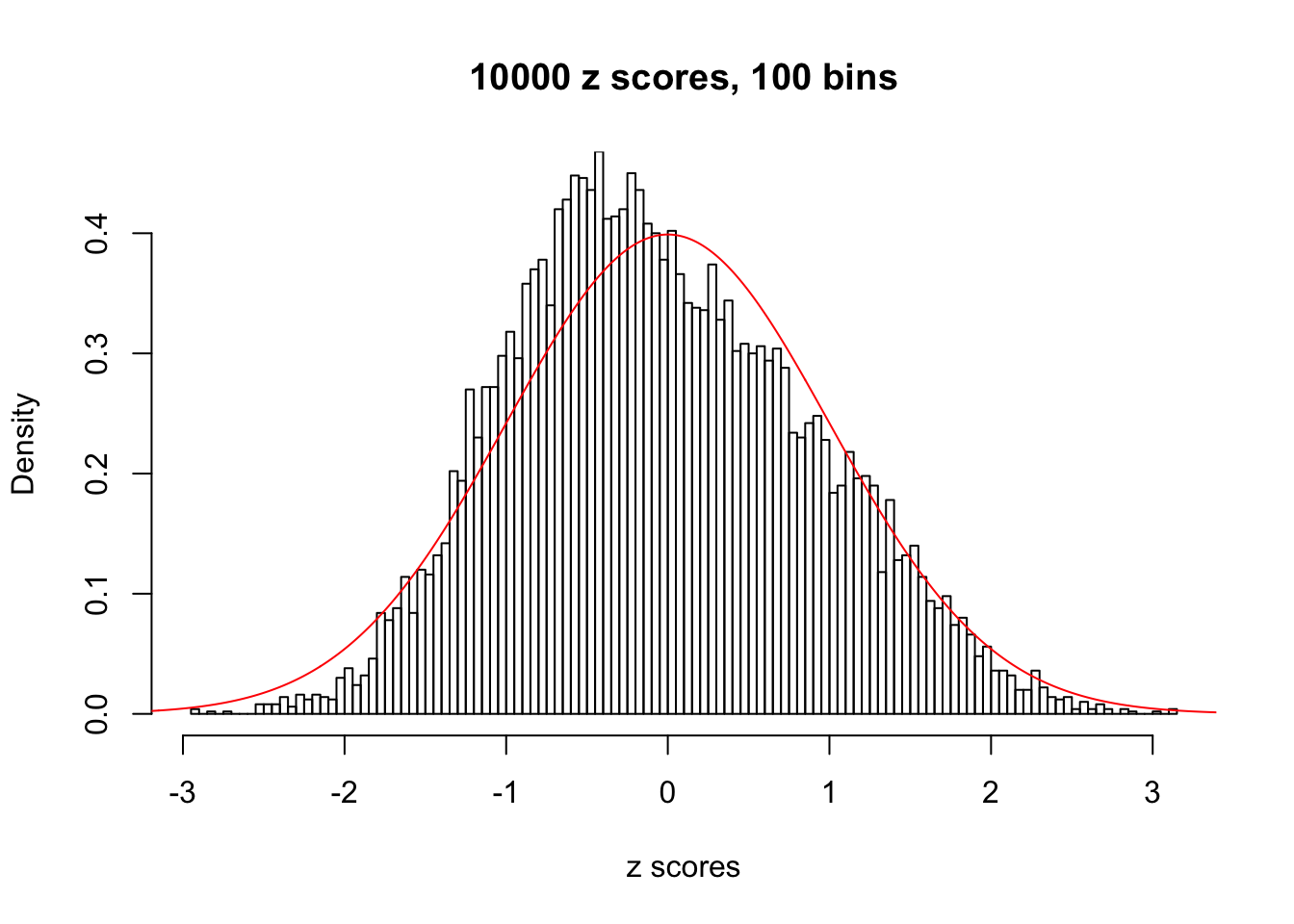
Data Set 383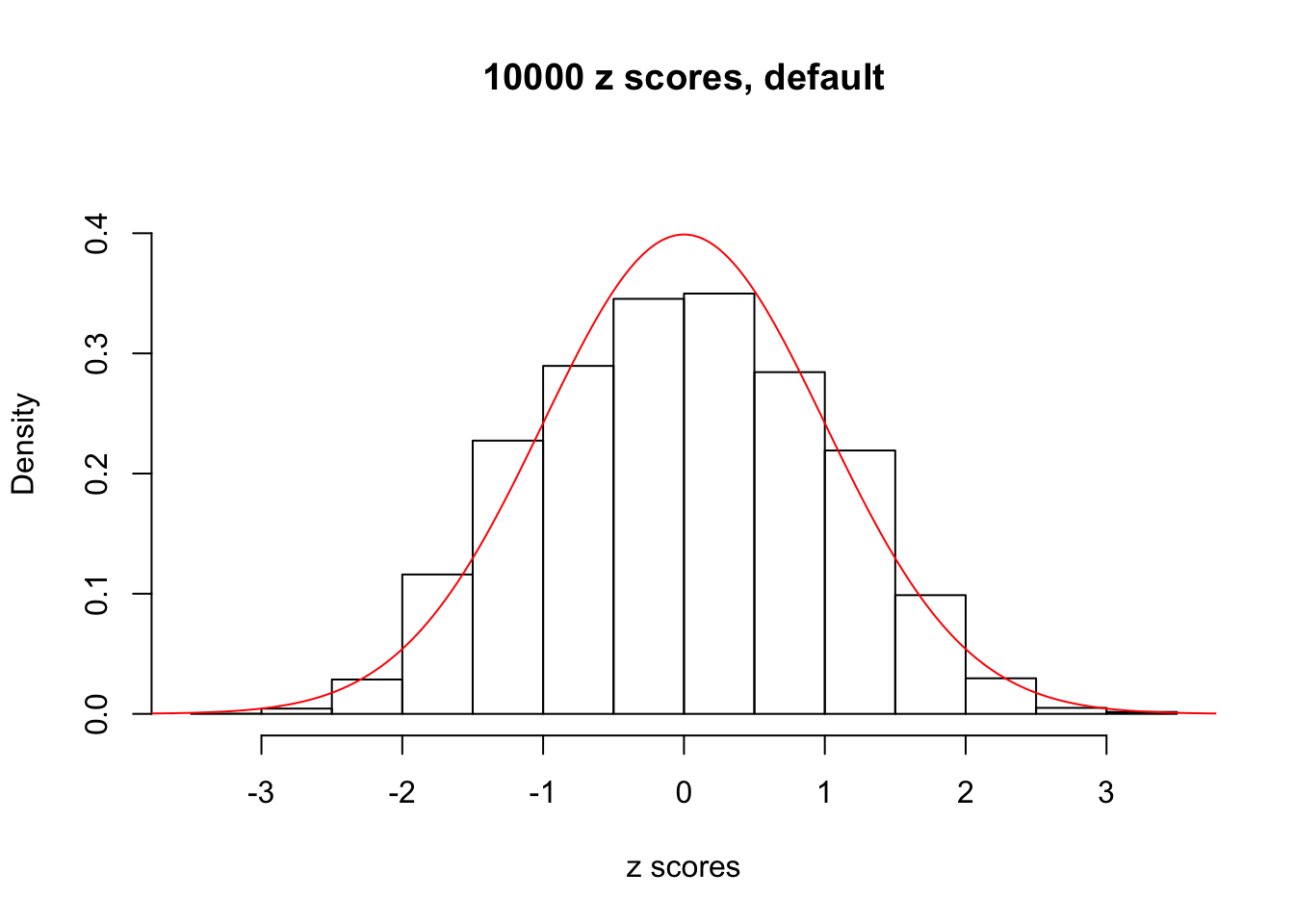
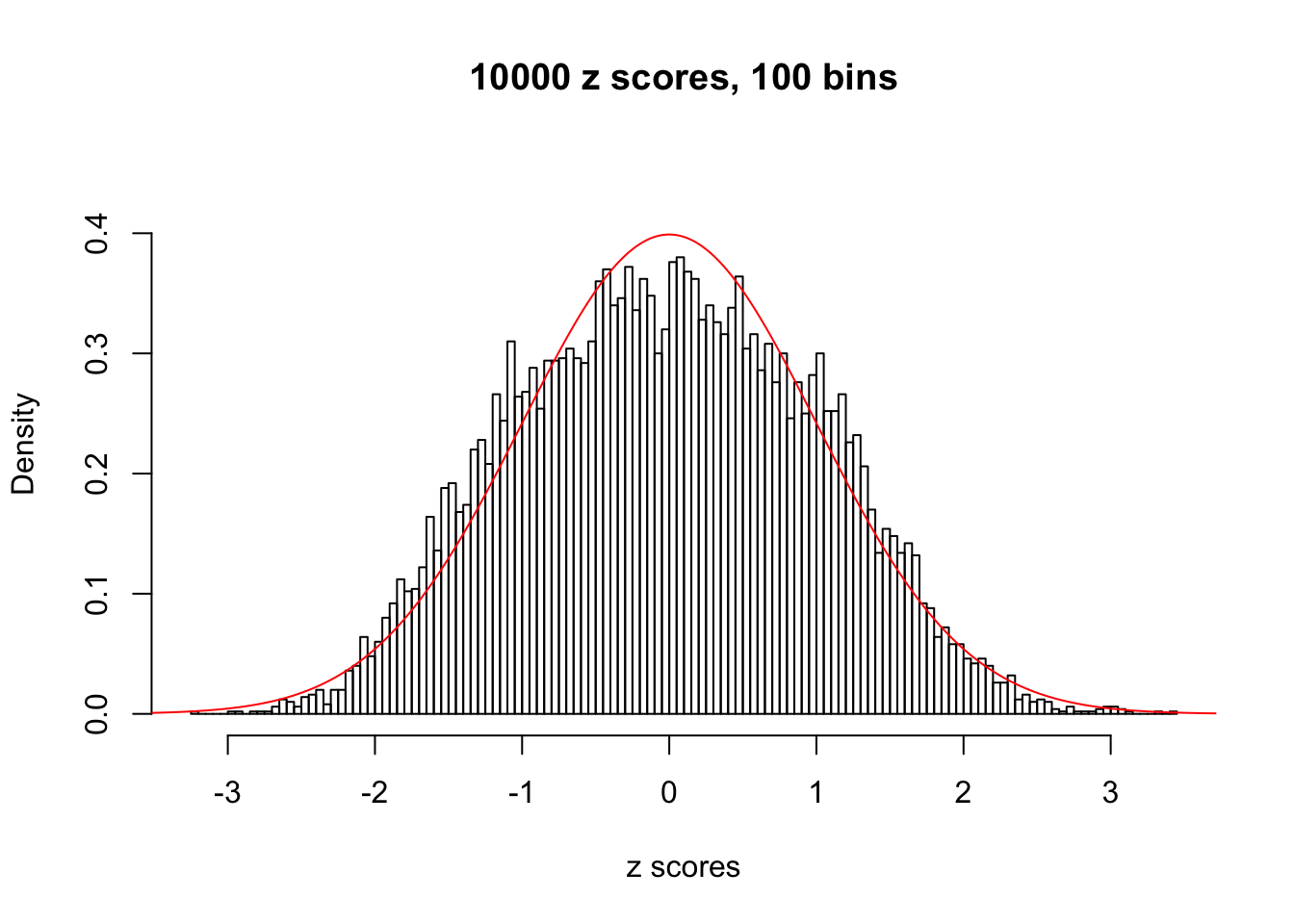
Data Set 412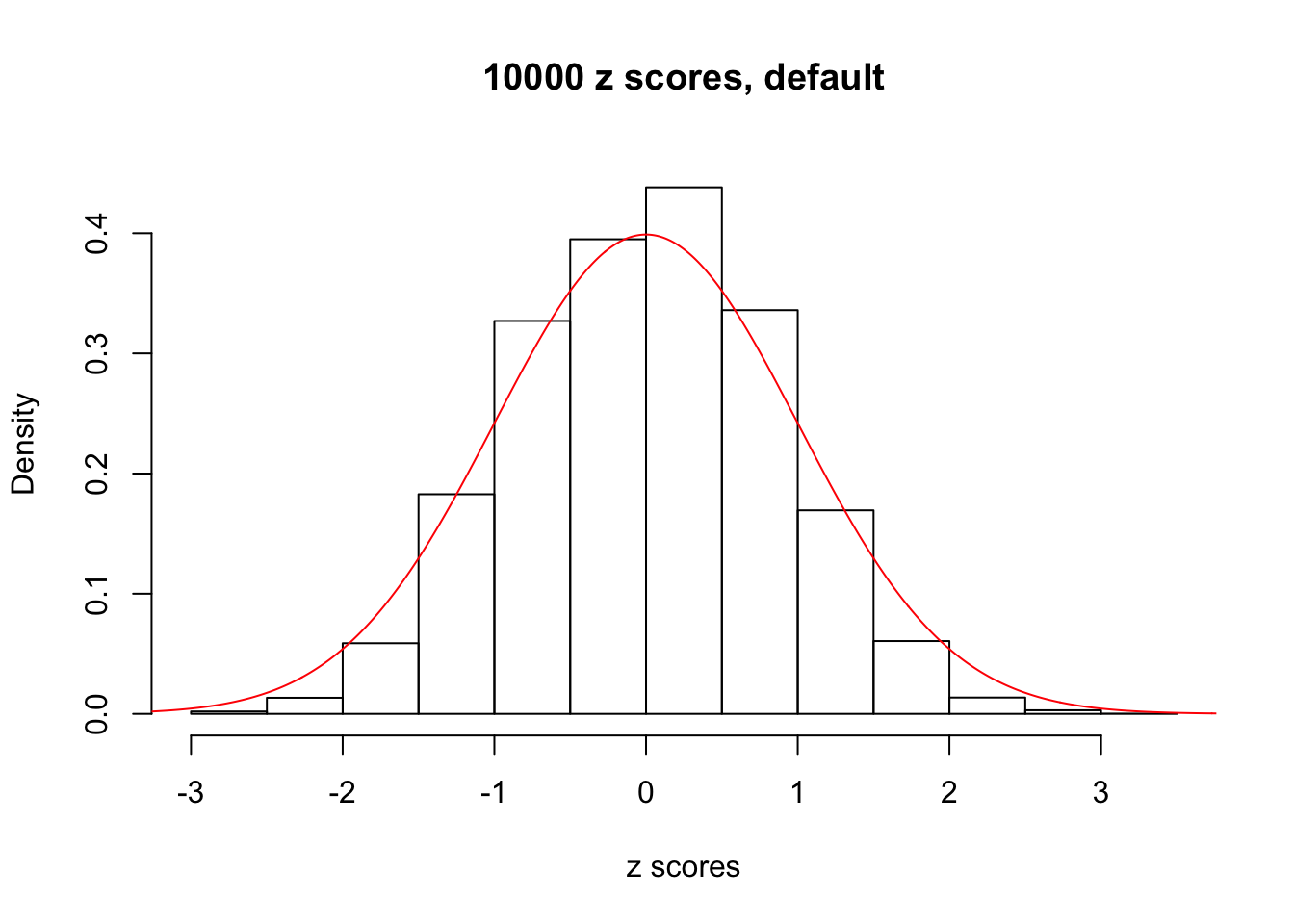
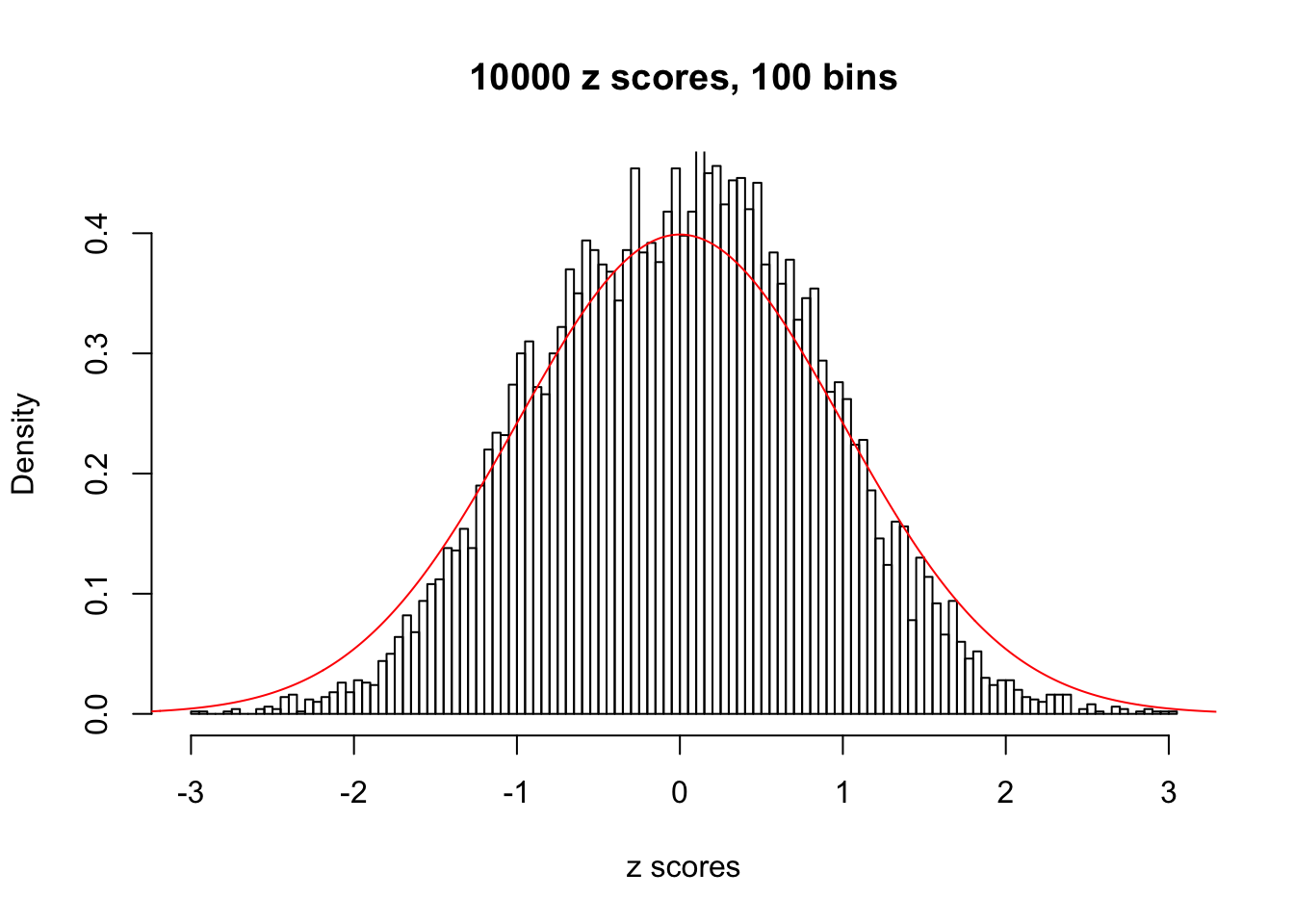
Data Set 492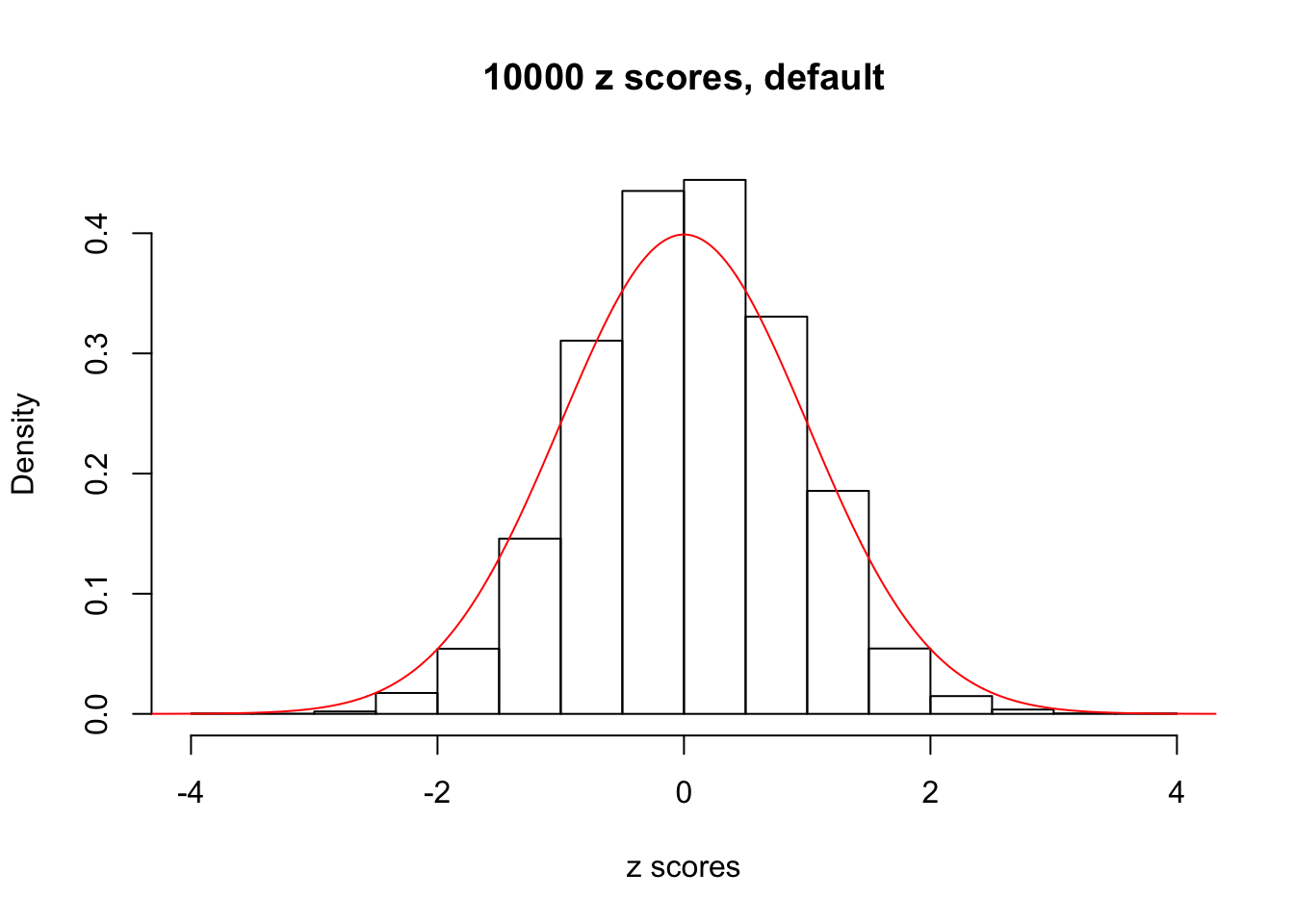
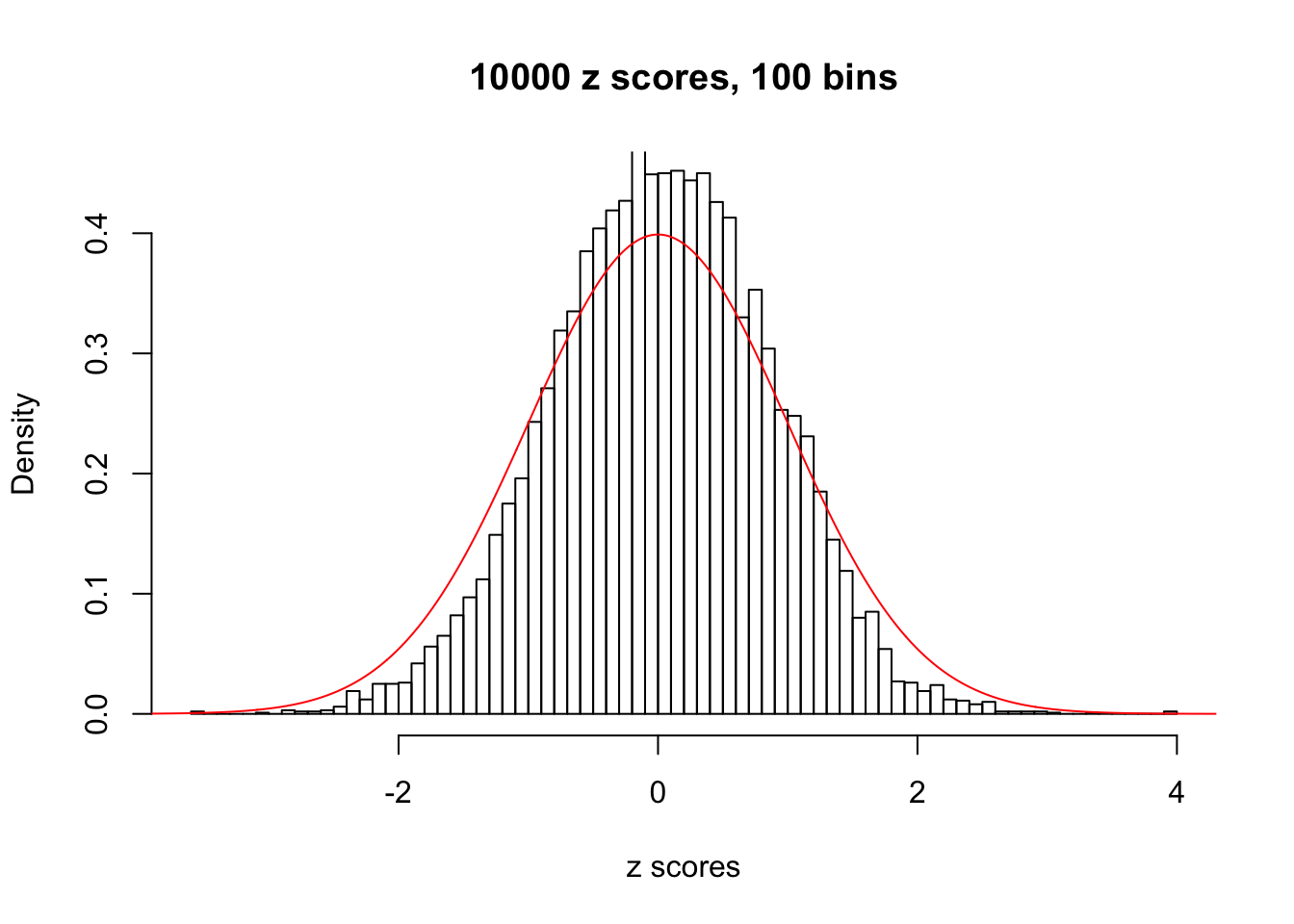
Data Set 574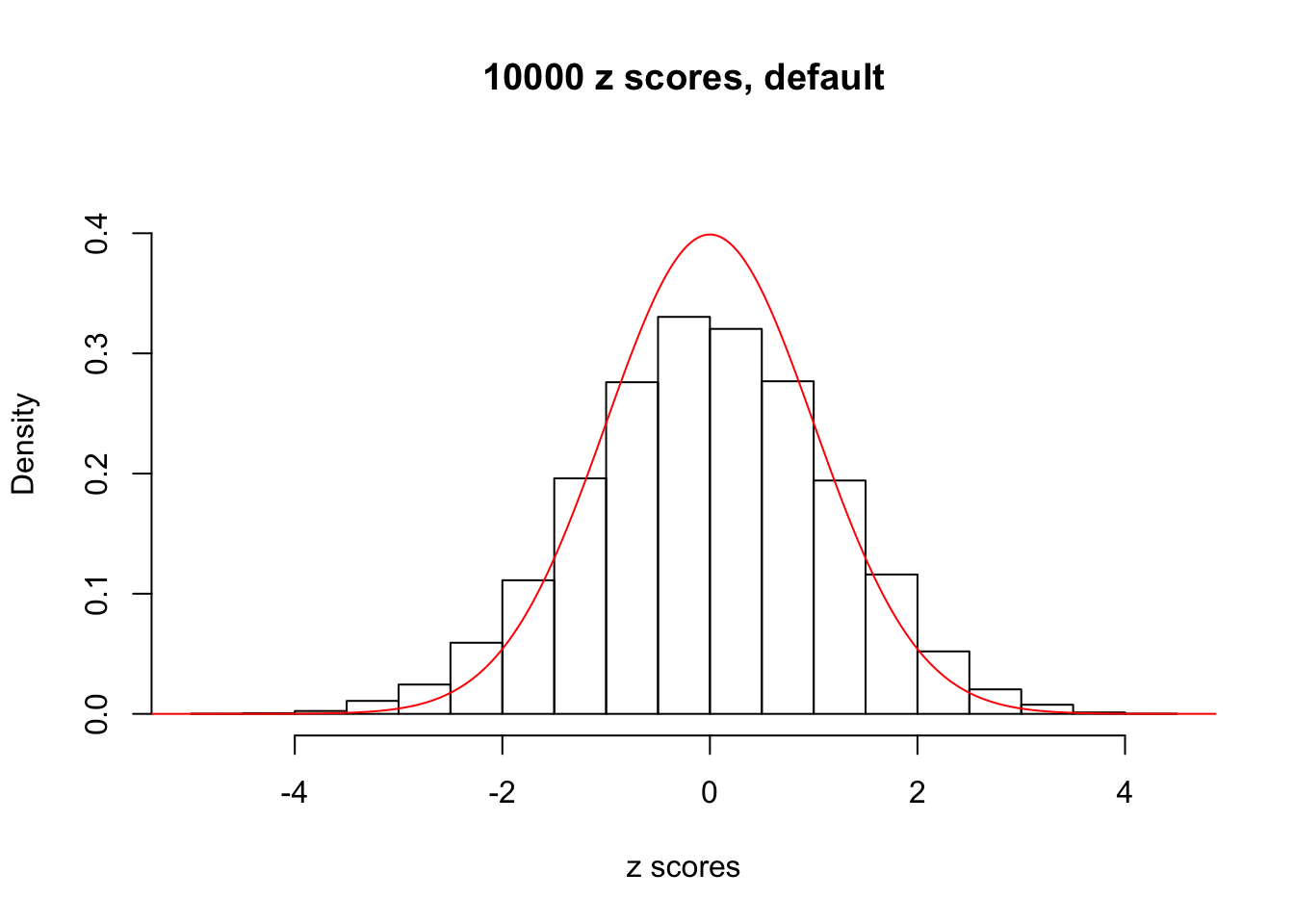
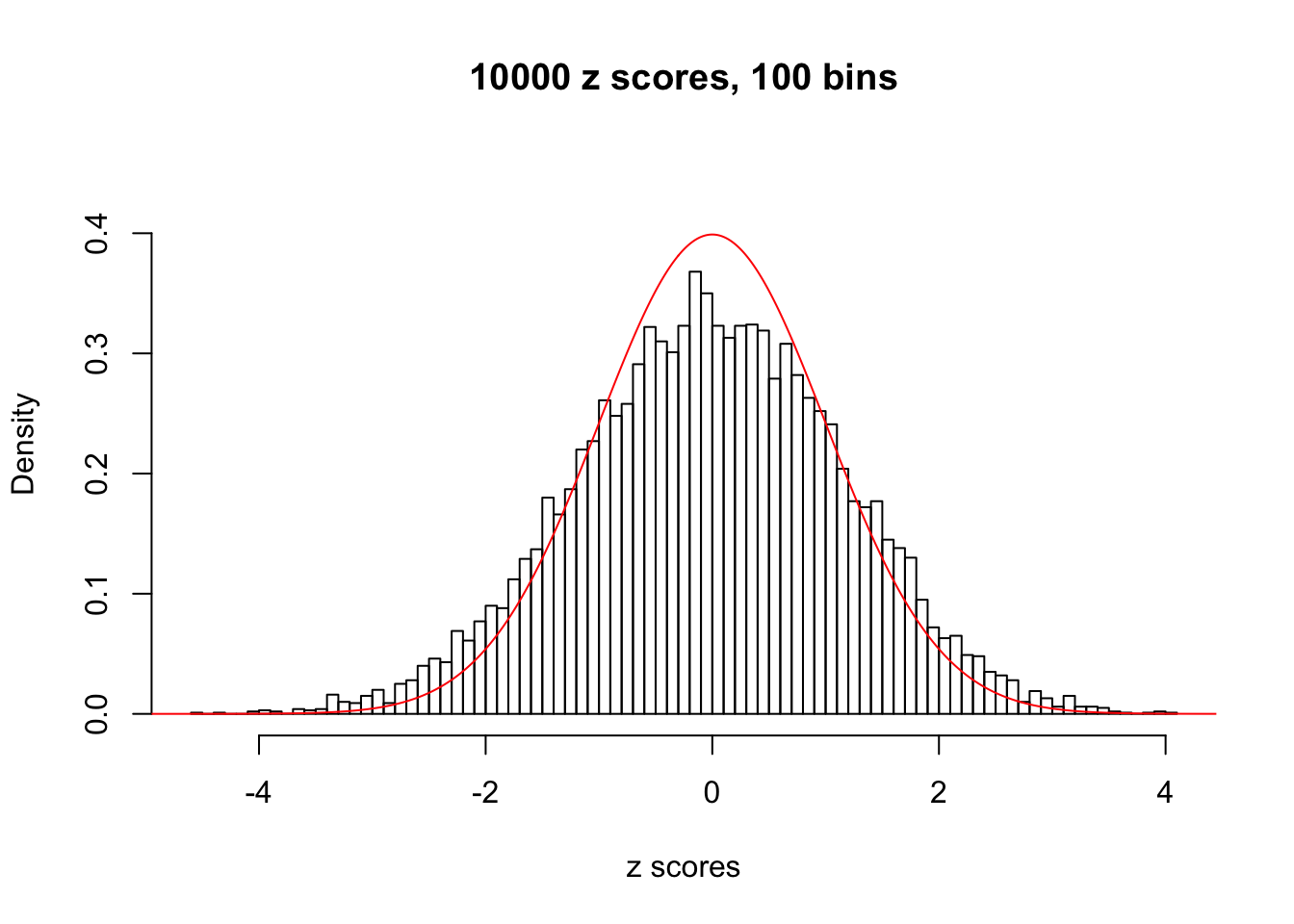
Data Set 588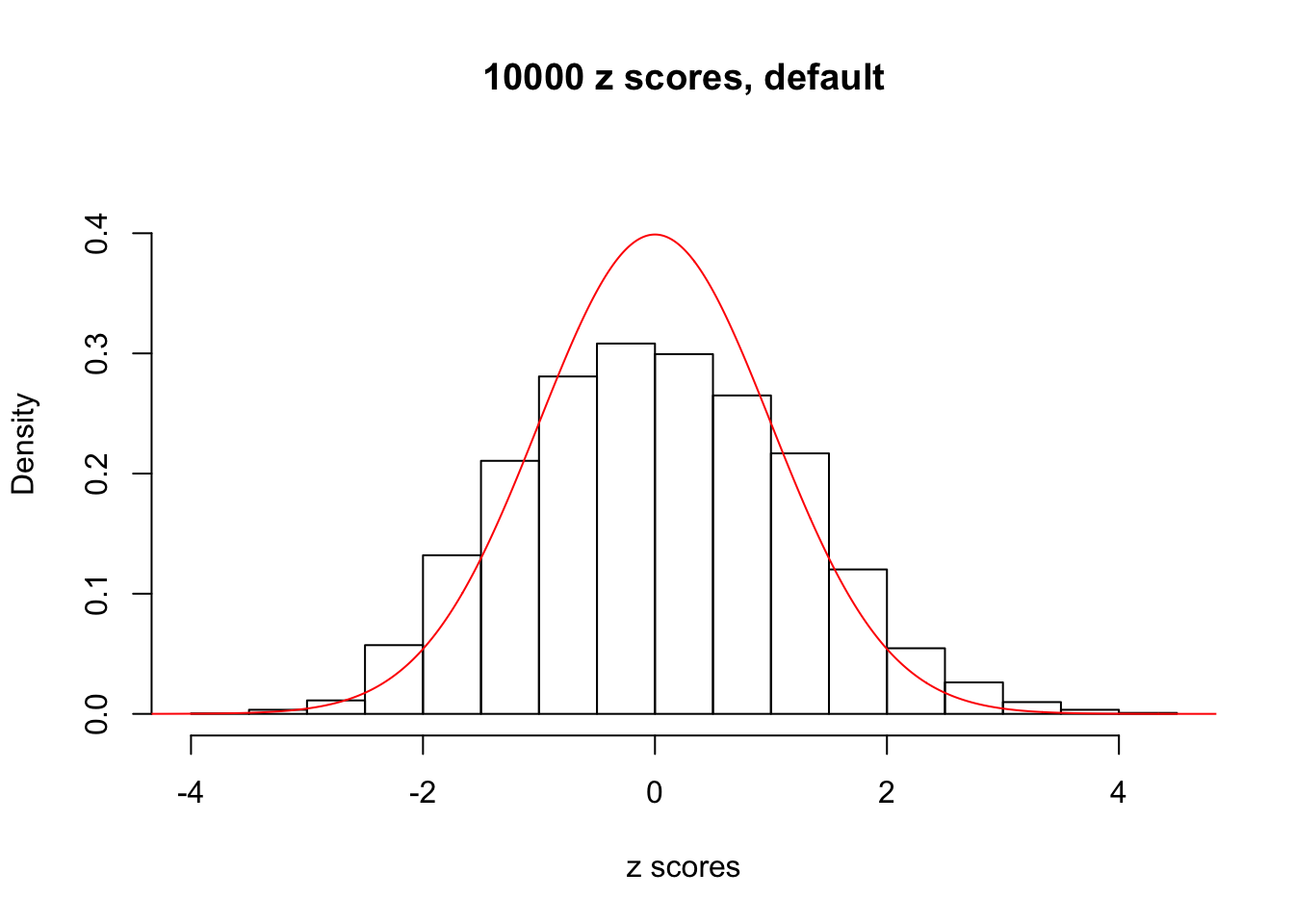
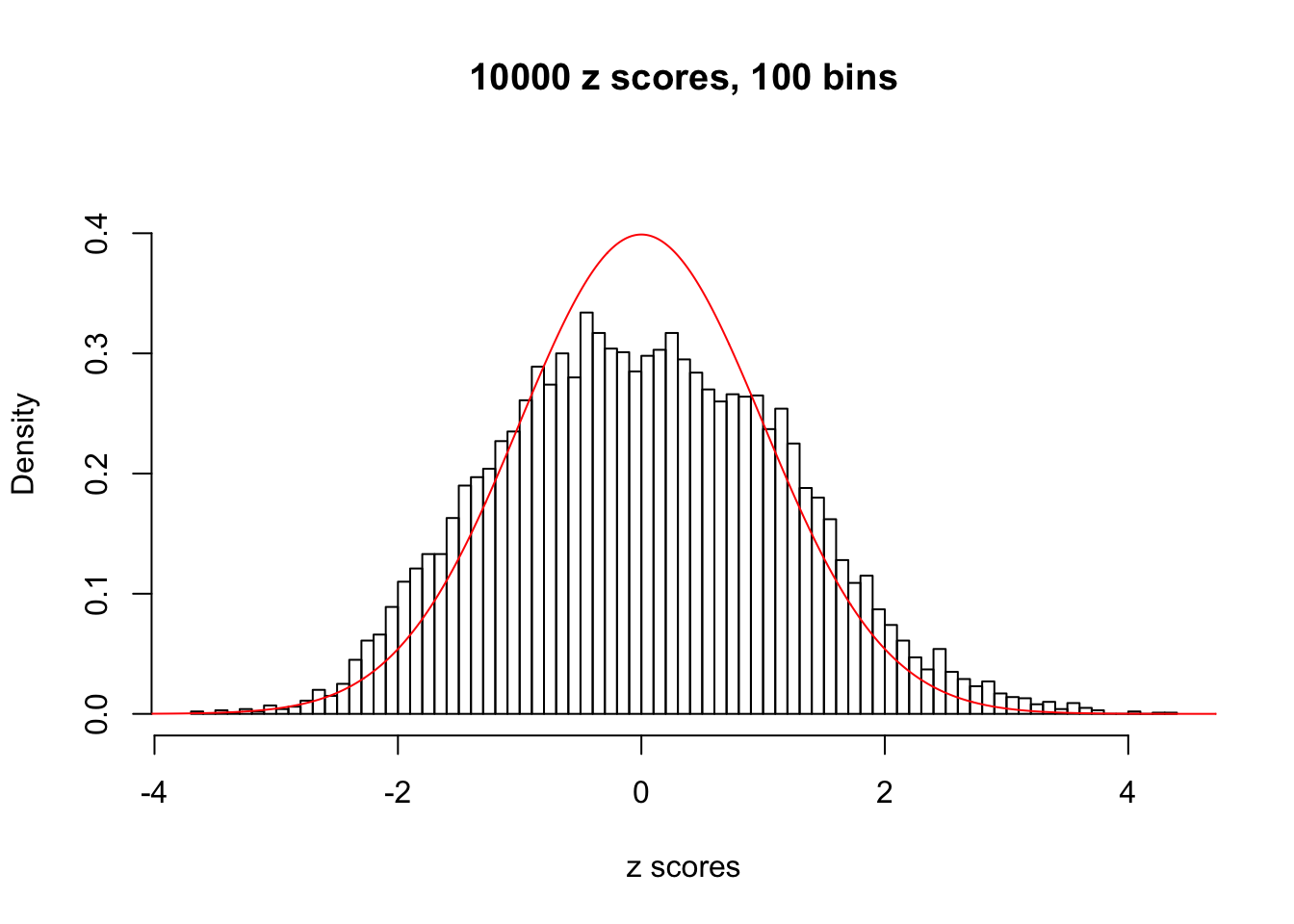
Data Set 654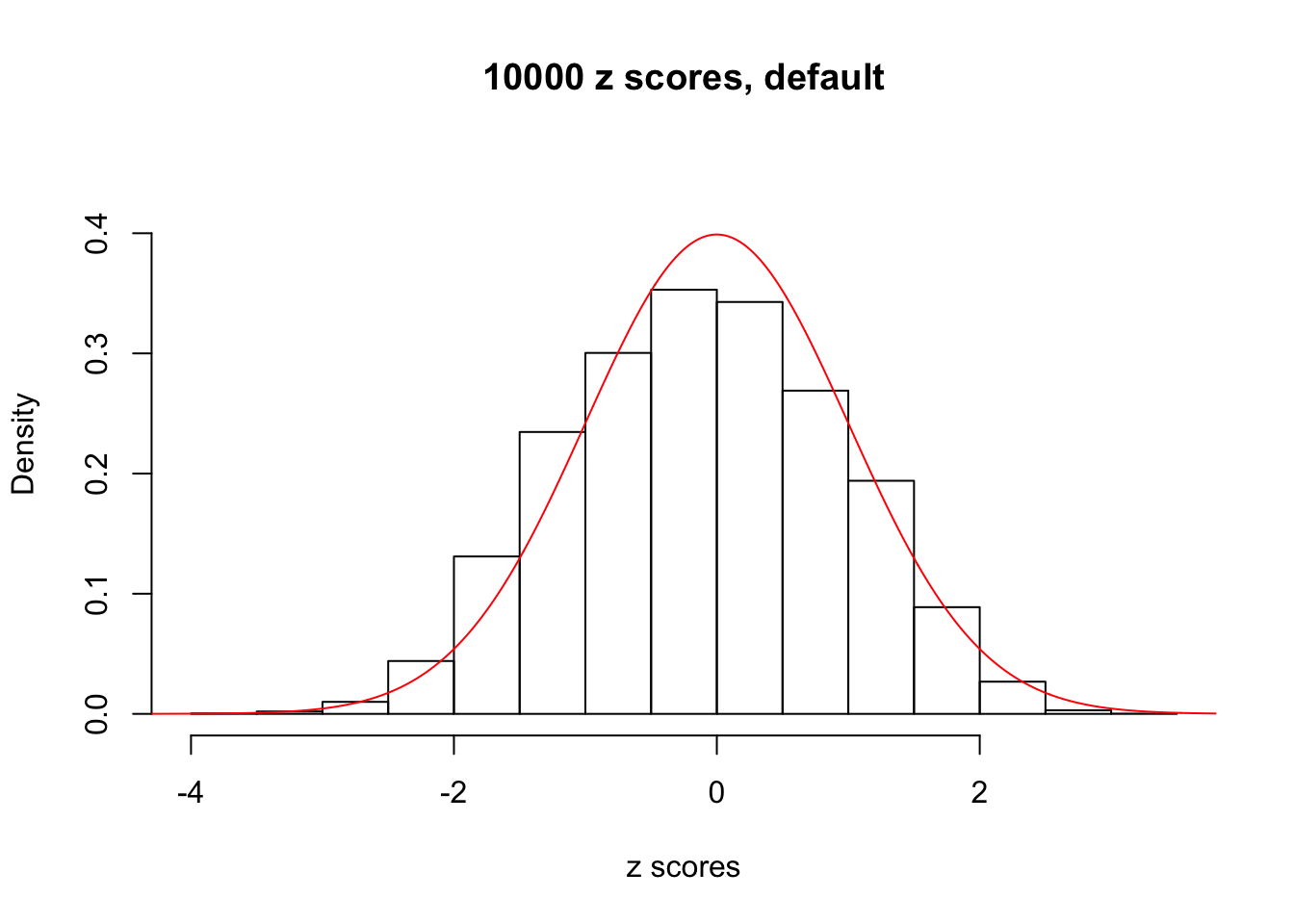
Data Set 688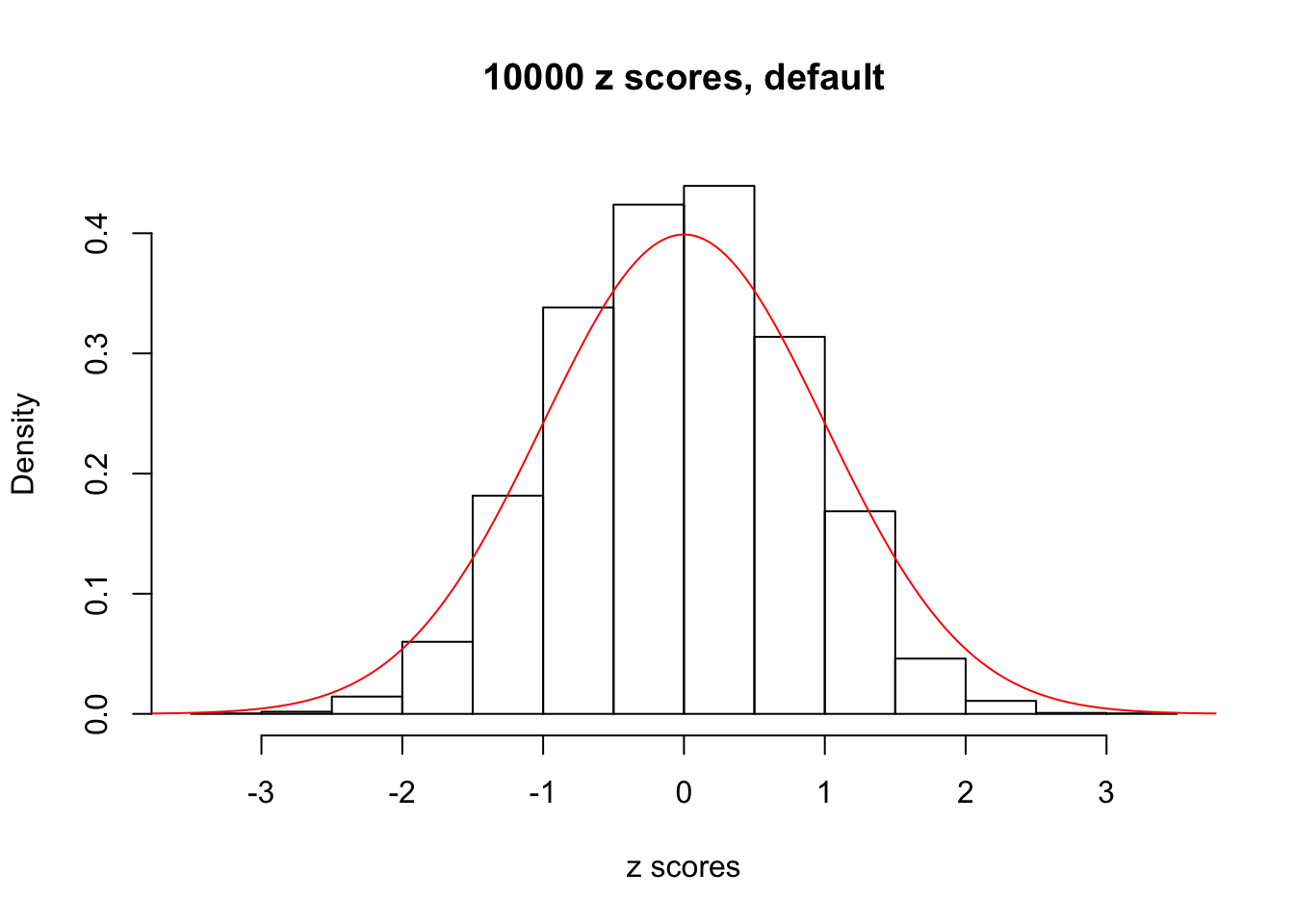
Data Set 693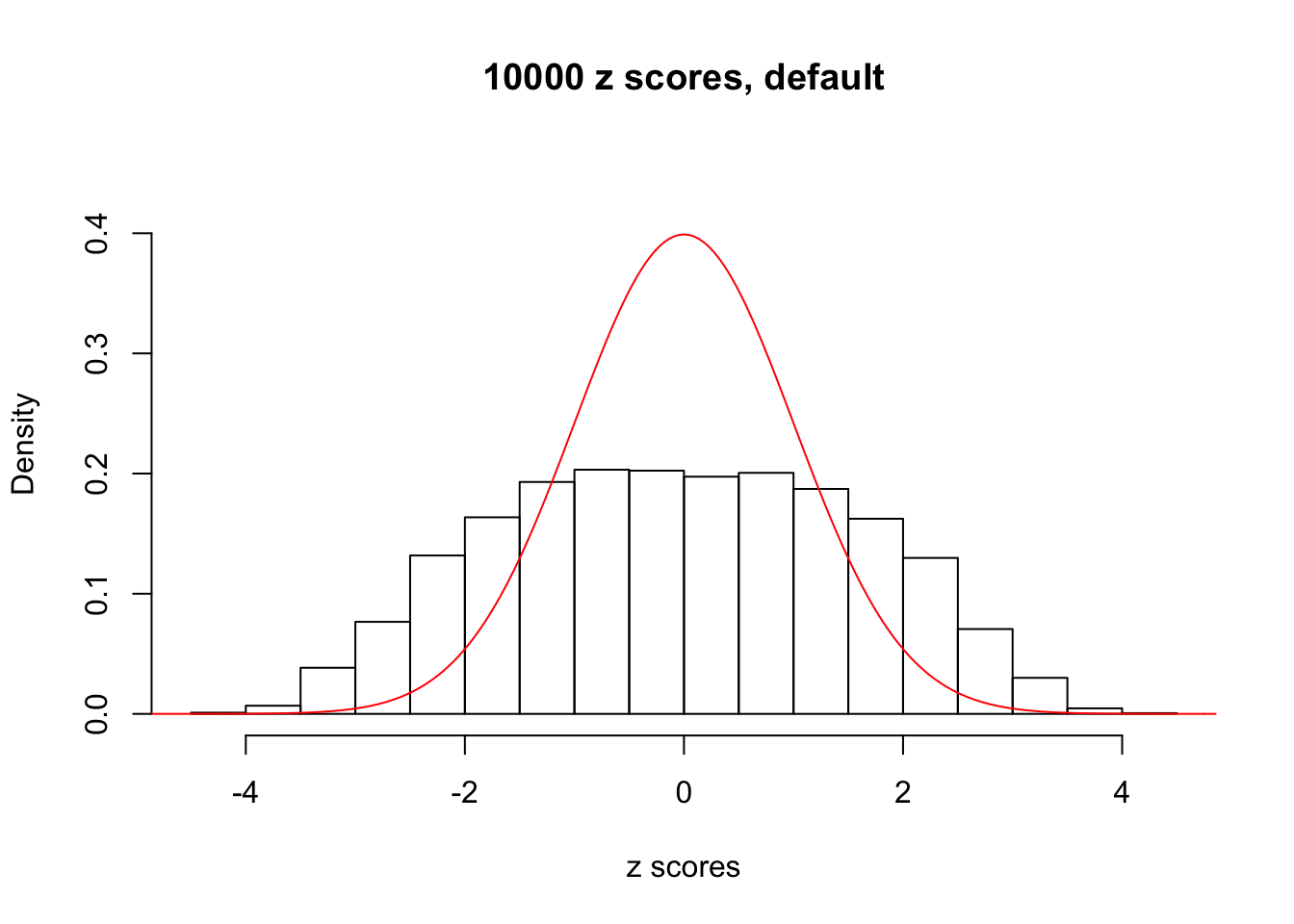
Data Set 726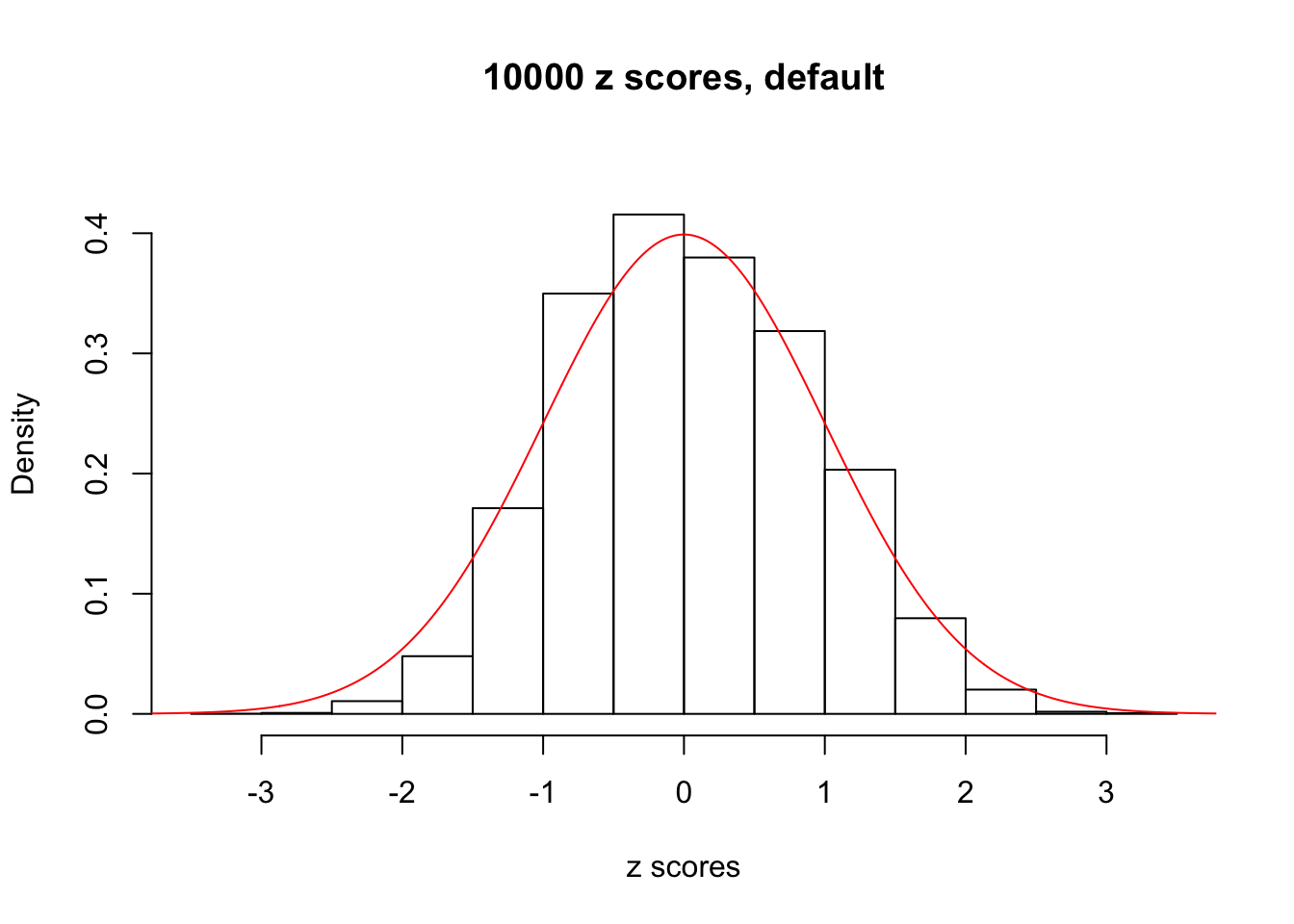
Data Set 853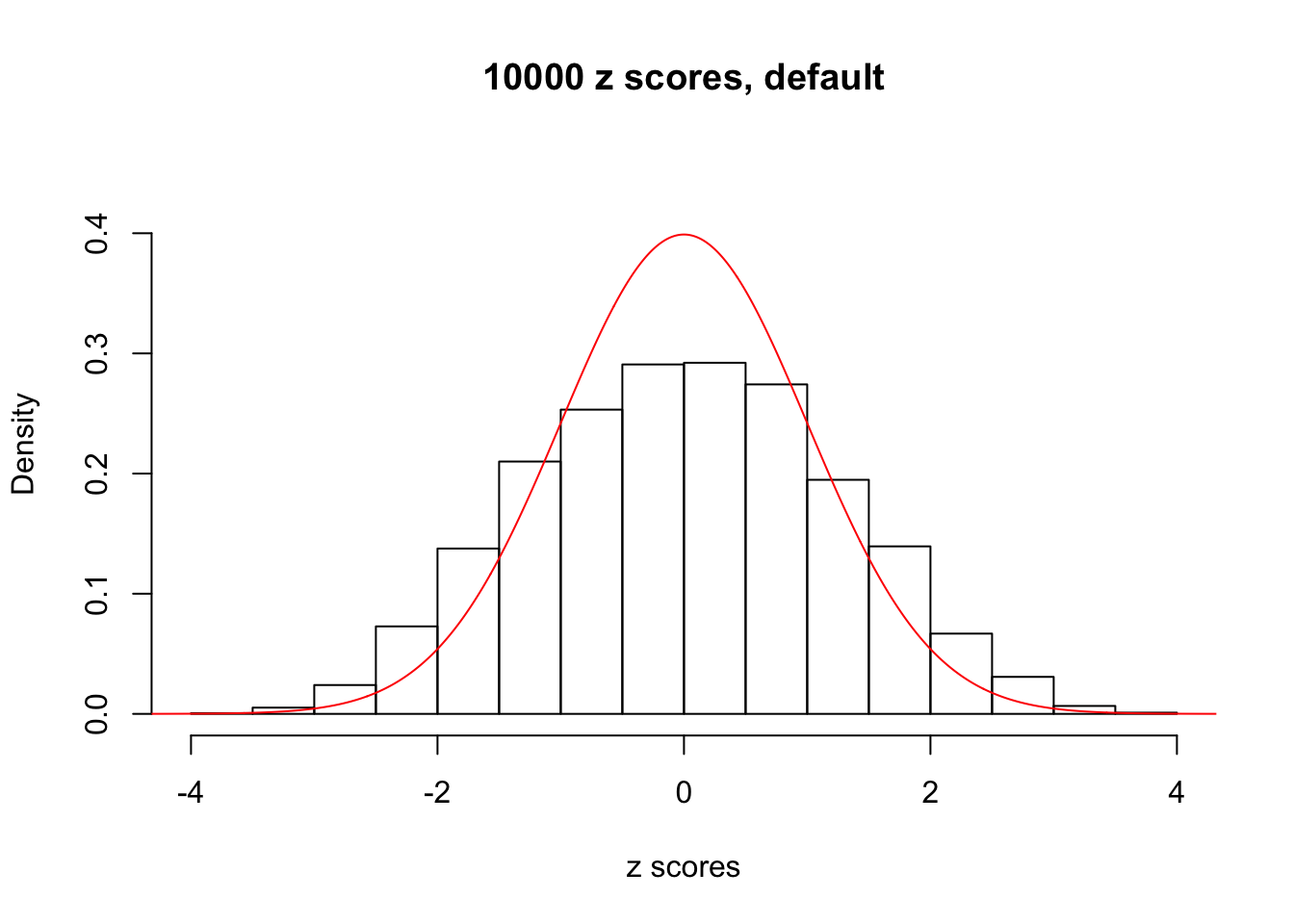
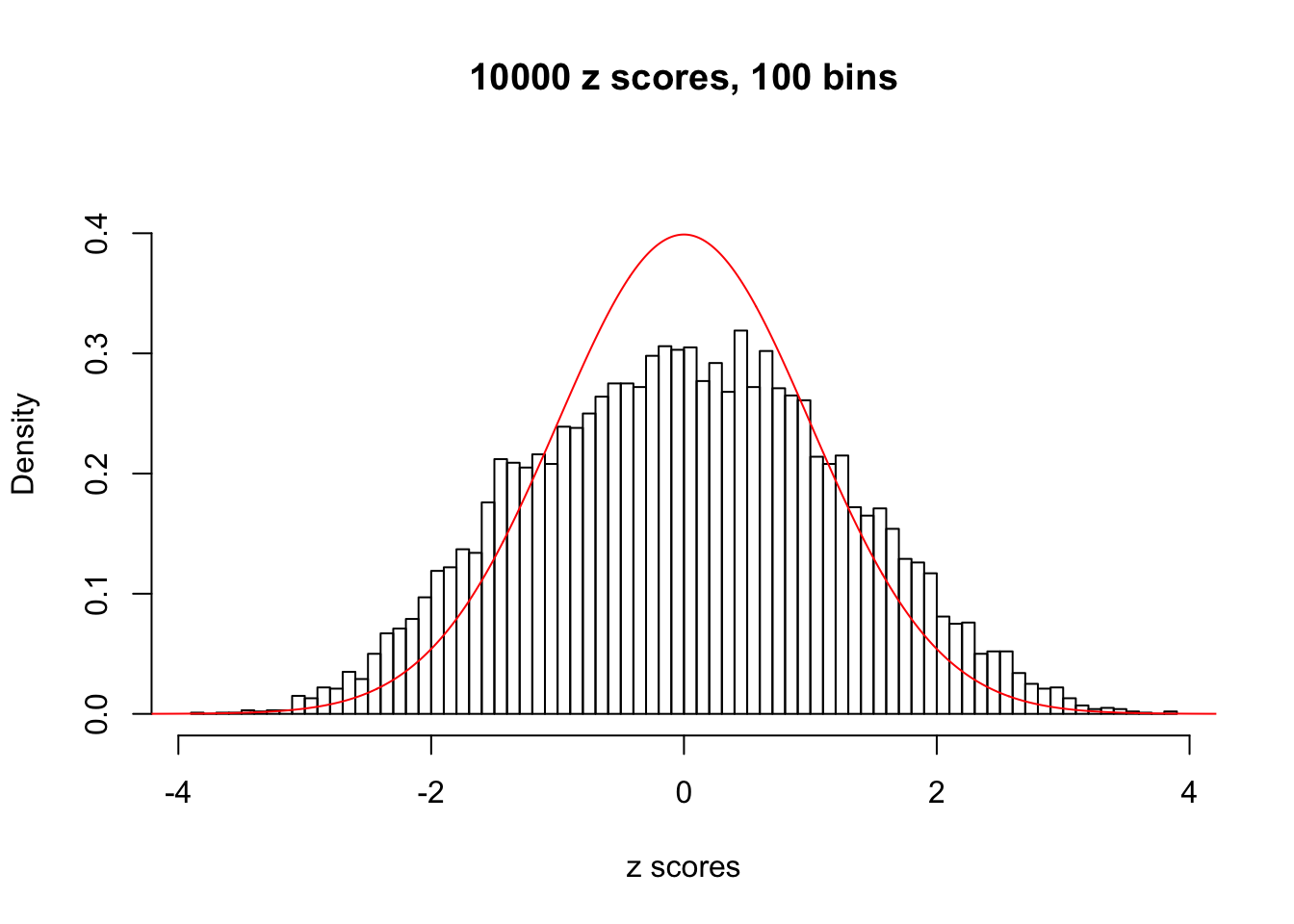
Data Set 855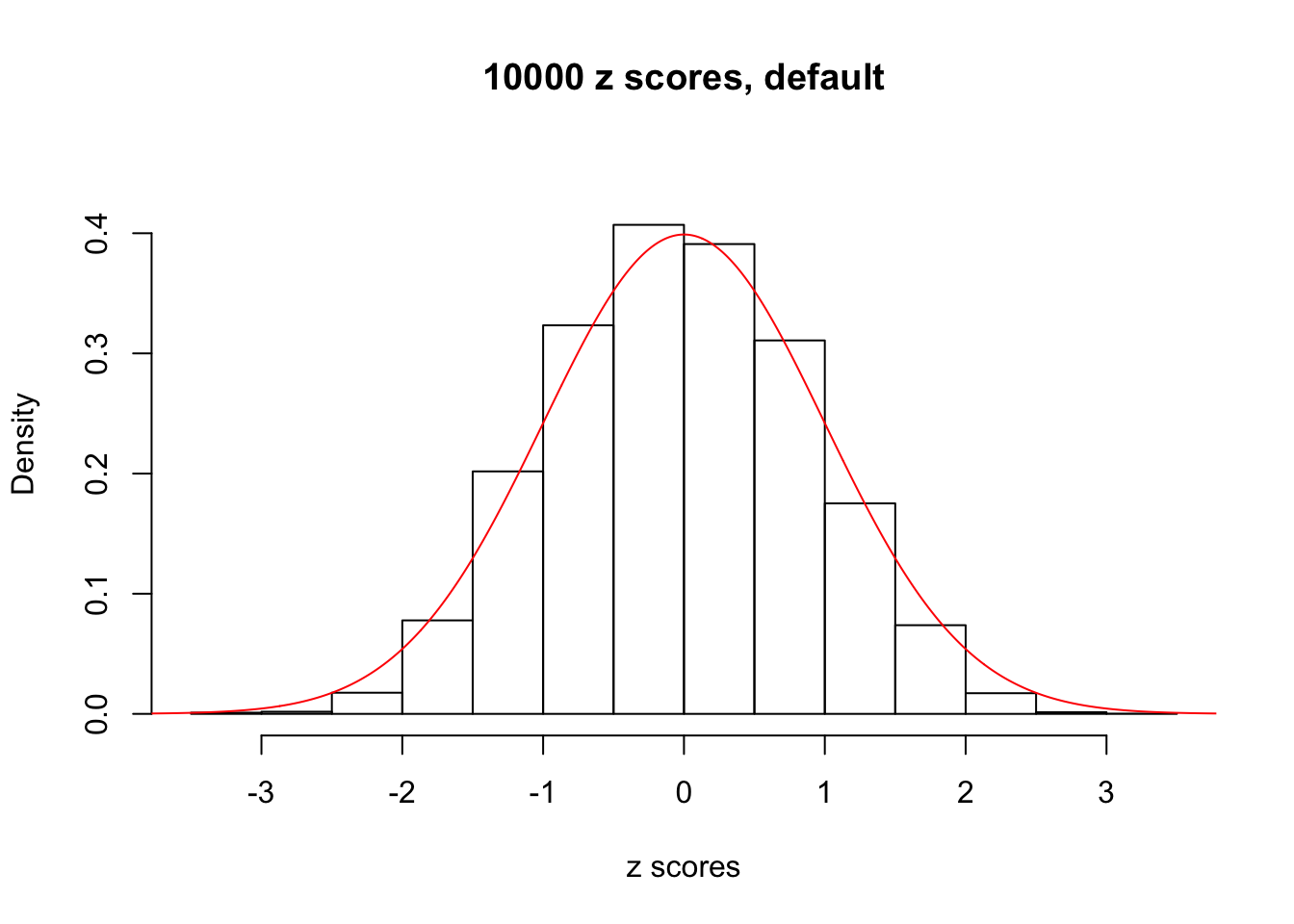
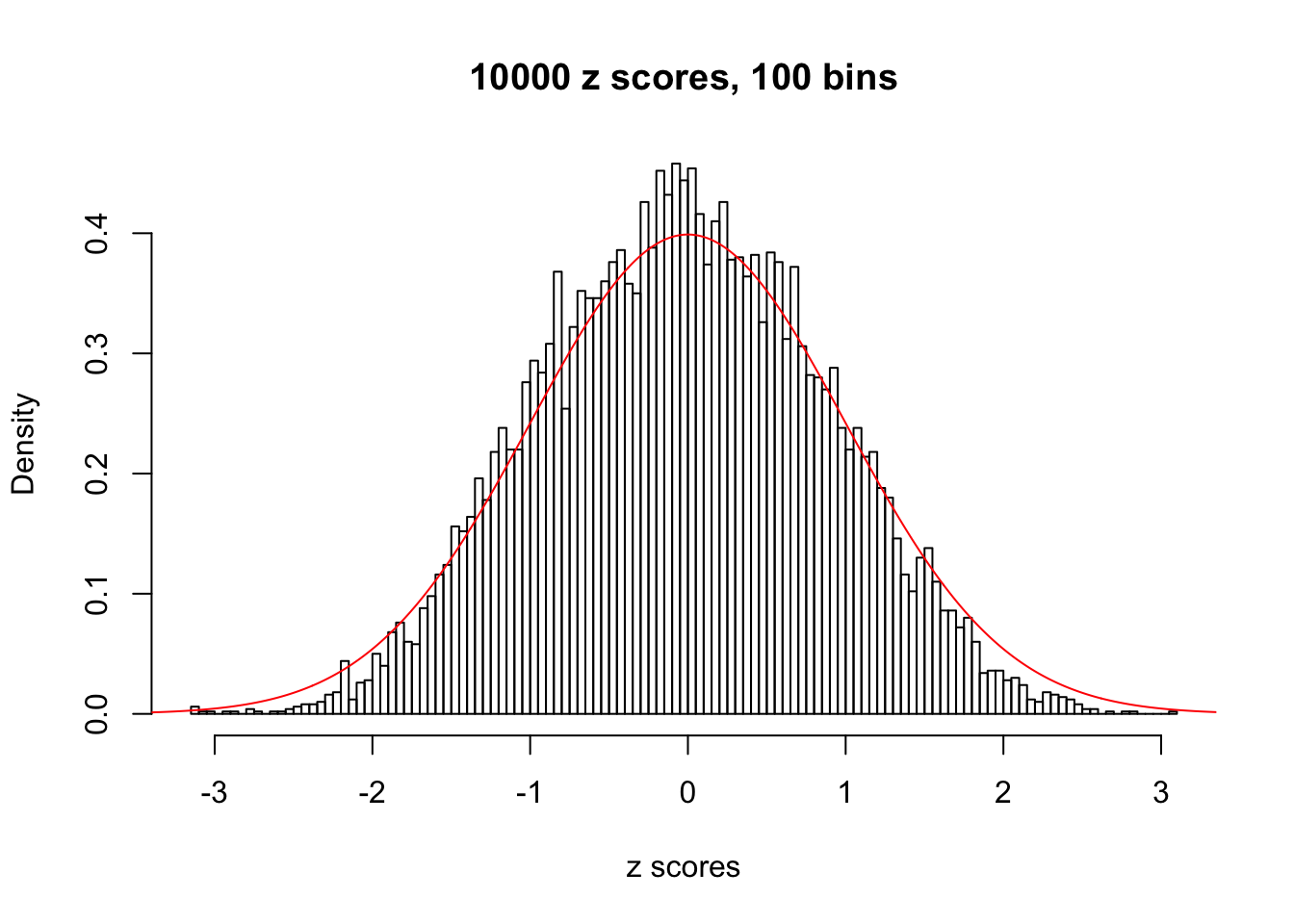
Data Set 942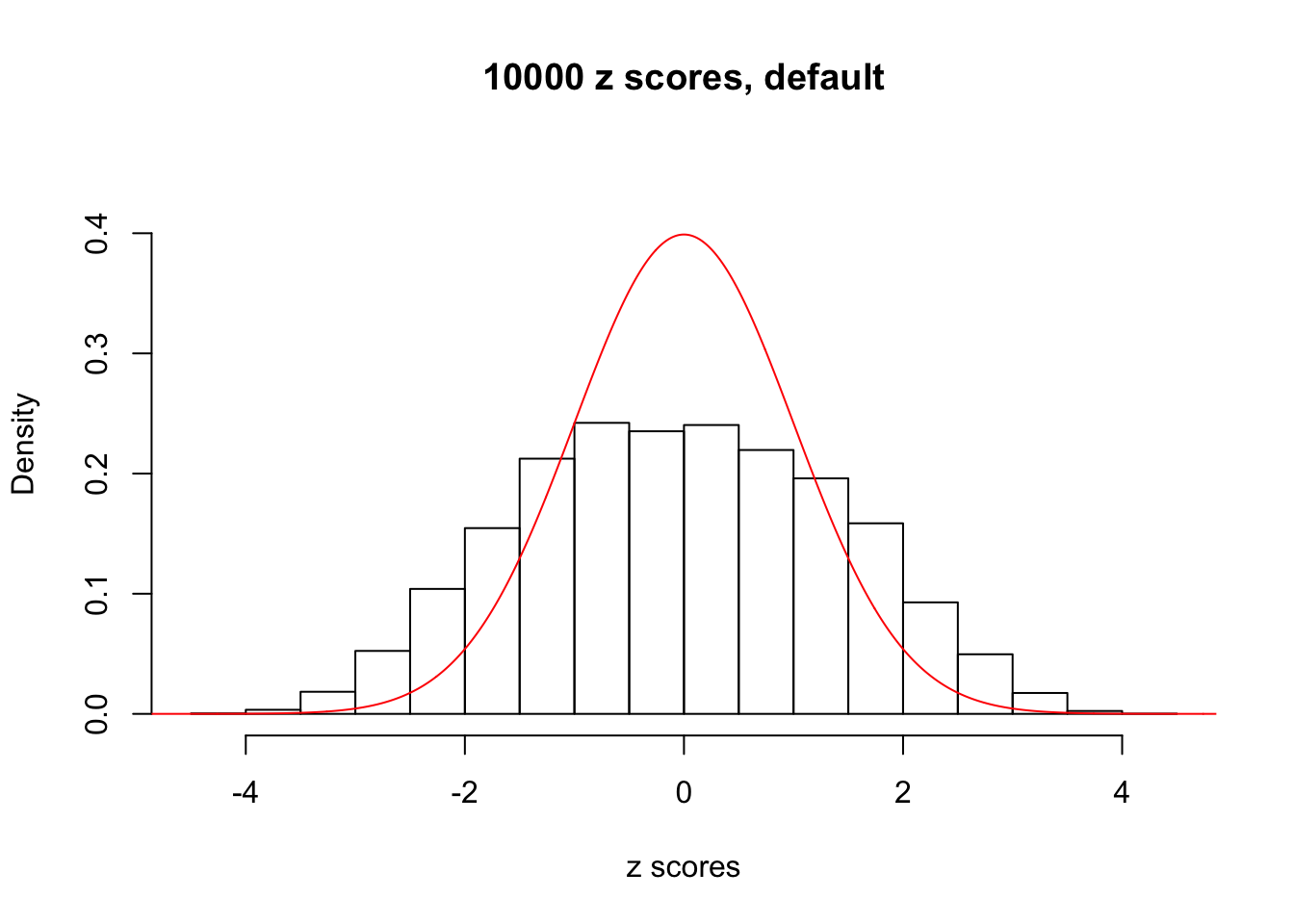
Data Set 993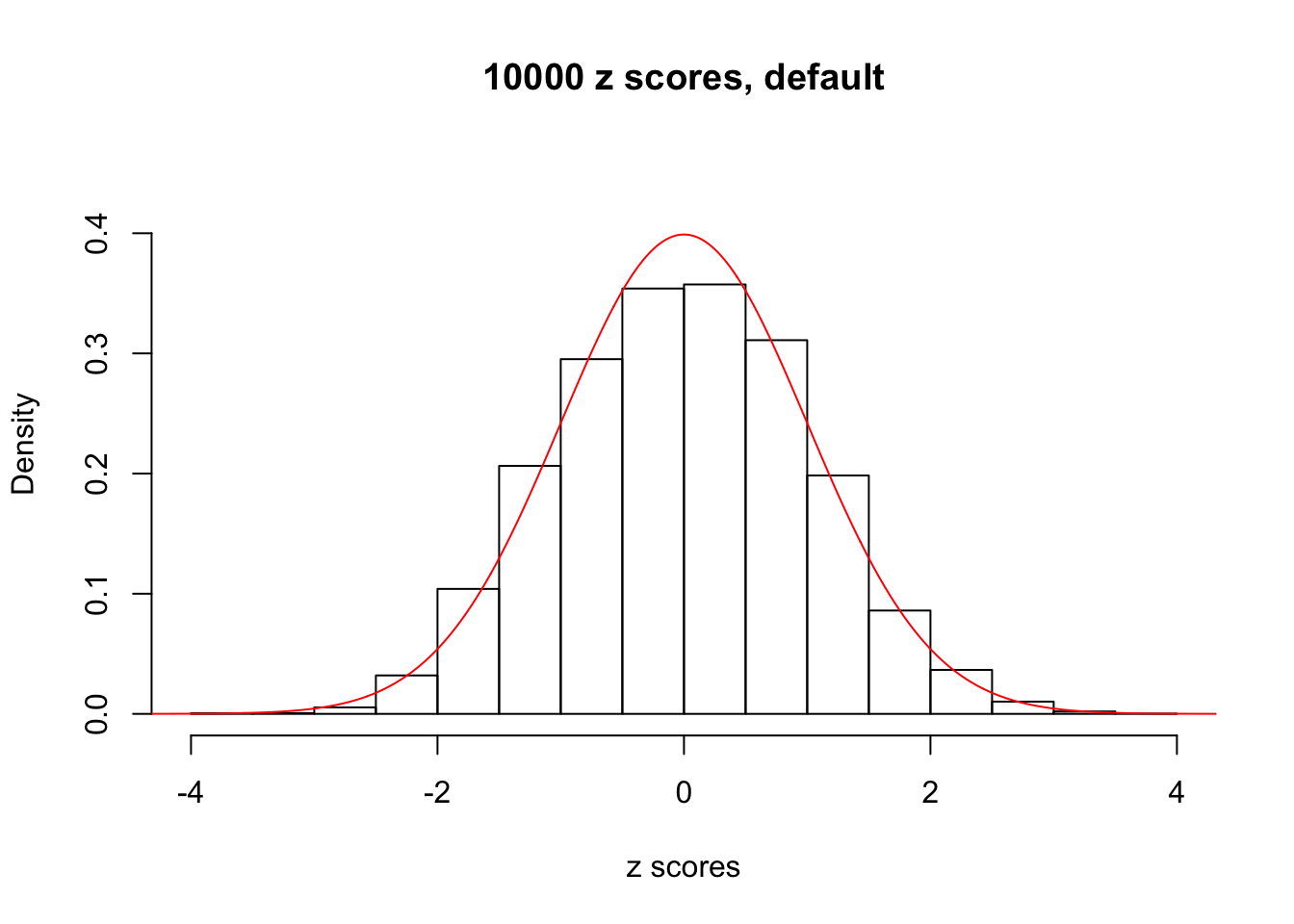
Session Information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_3.4.3 backports_1.1.2 magrittr_1.5 rprojroot_1.3-1
[5] tools_3.4.3 htmltools_0.3.6 yaml_2.1.16 Rcpp_0.12.14
[9] stringi_1.1.6 rmarkdown_1.8 knitr_1.17 git2r_0.20.0
[13] stringr_1.2.0 digest_0.6.13 workflowr_0.8.0 evaluate_0.10.1This R Markdown site was created with workflowr