Simulating Correlated \(N(0, 1)\) \(Z\) Scores: More Exotic
Lei Sun
2017-05-31
Last updated: 2017-12-21
Code version: 6e42447
Introduction
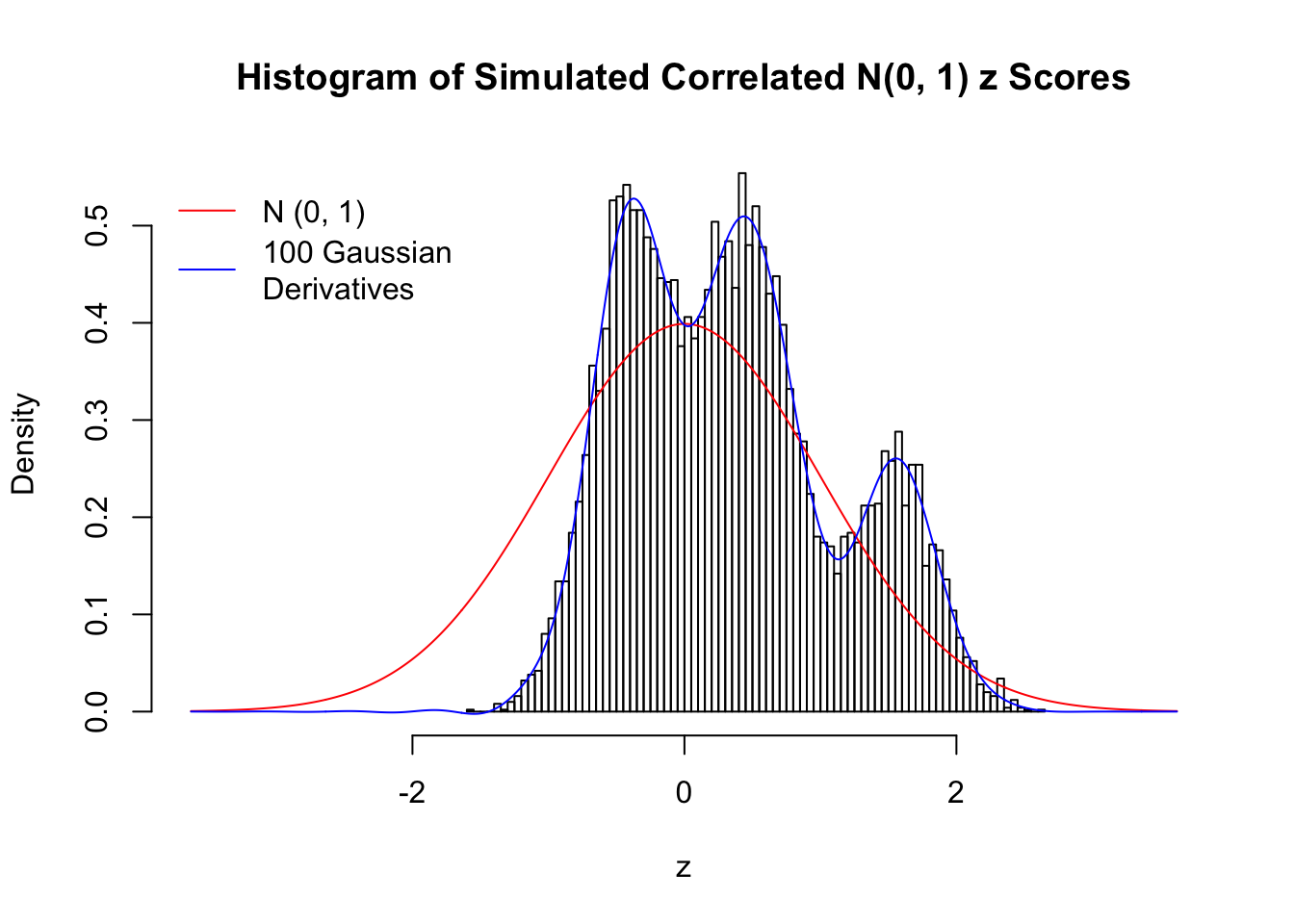
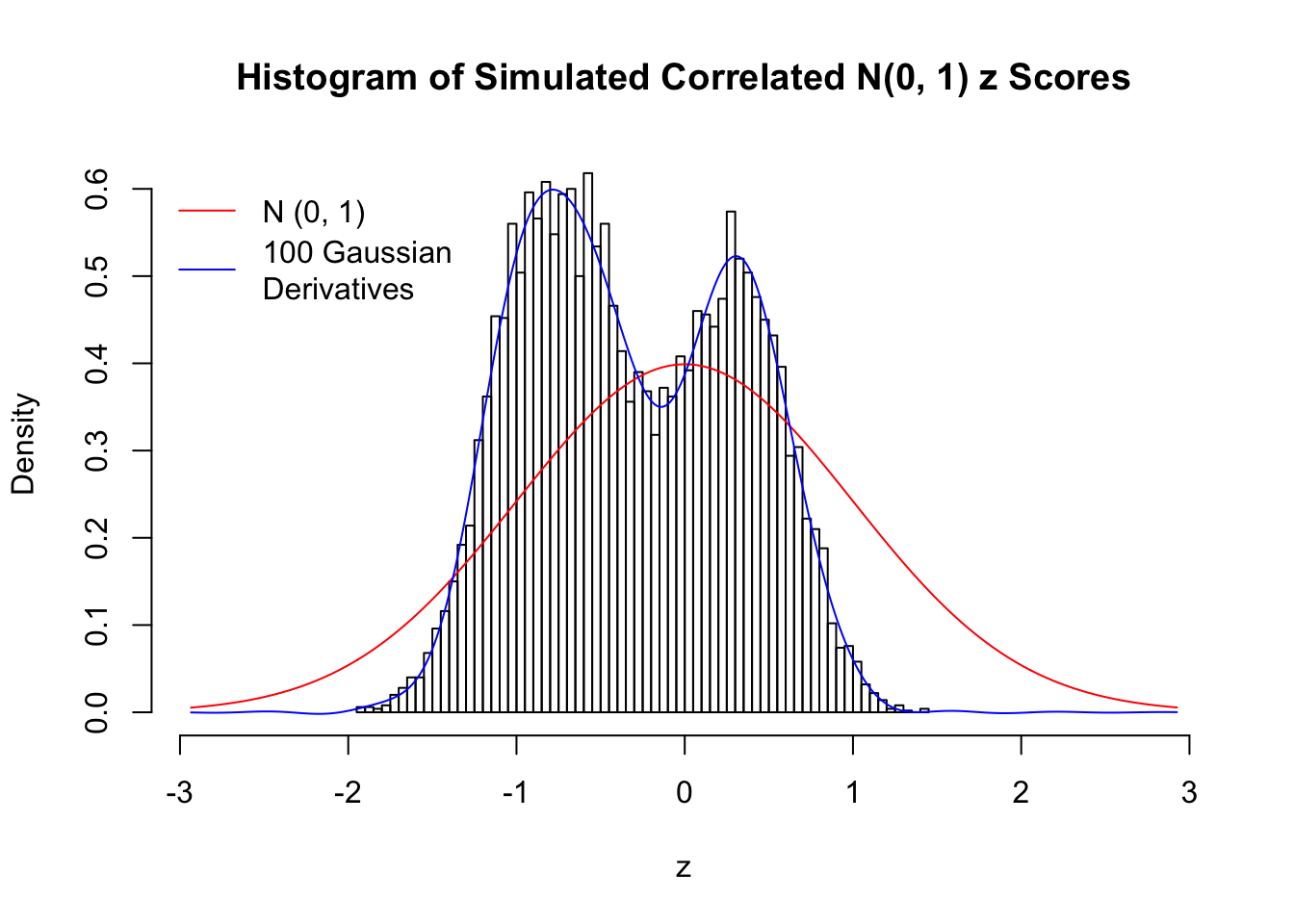
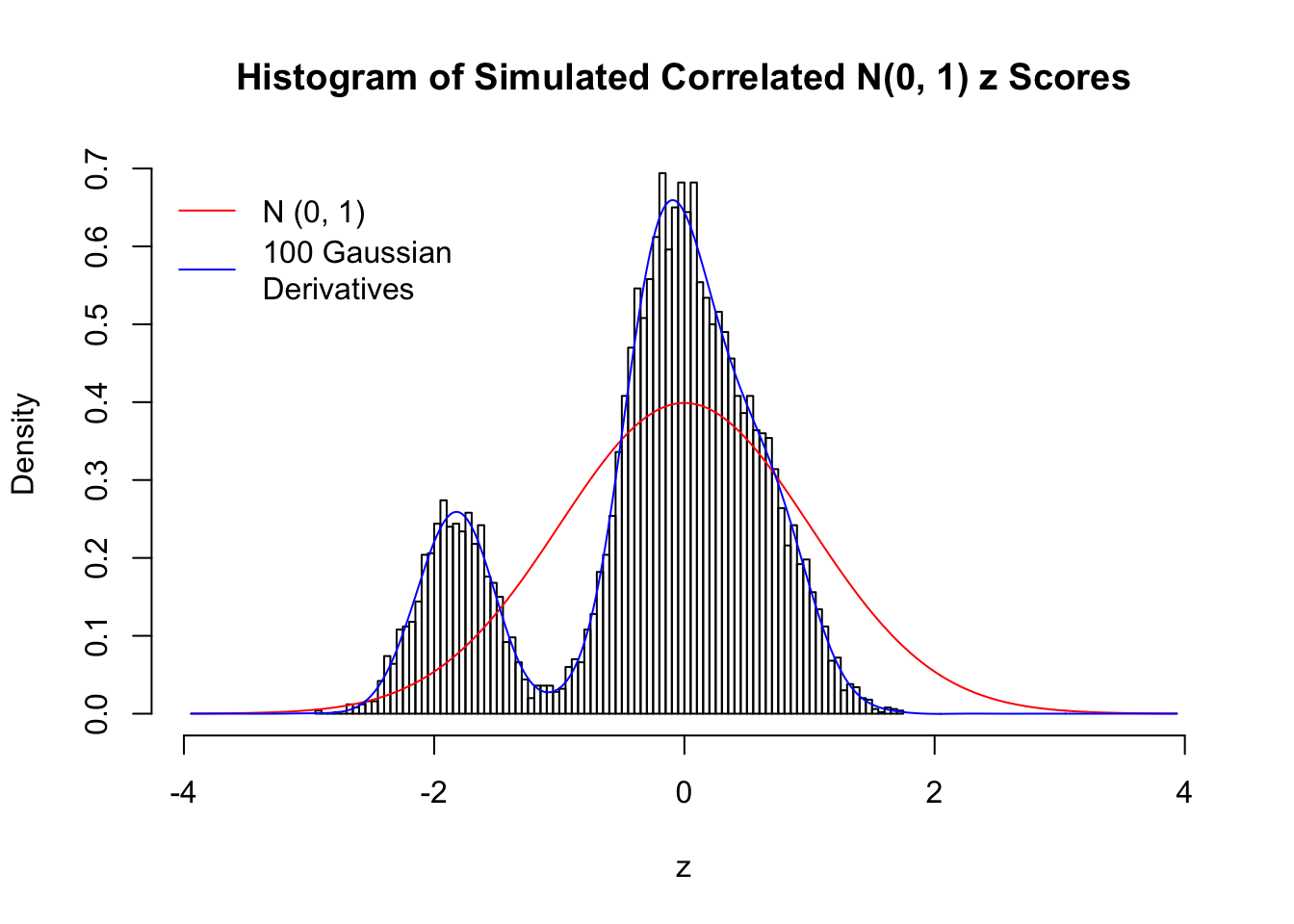
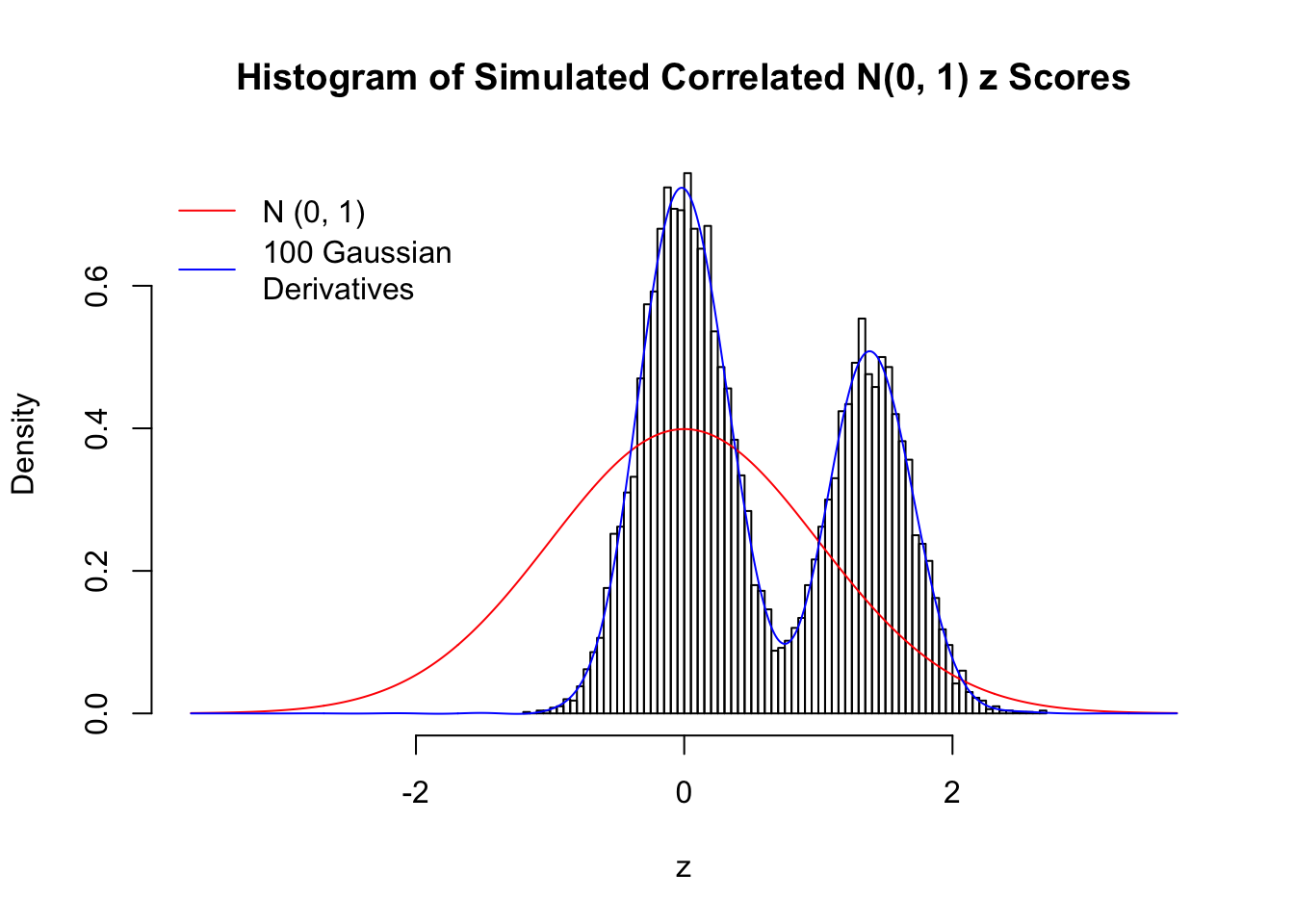
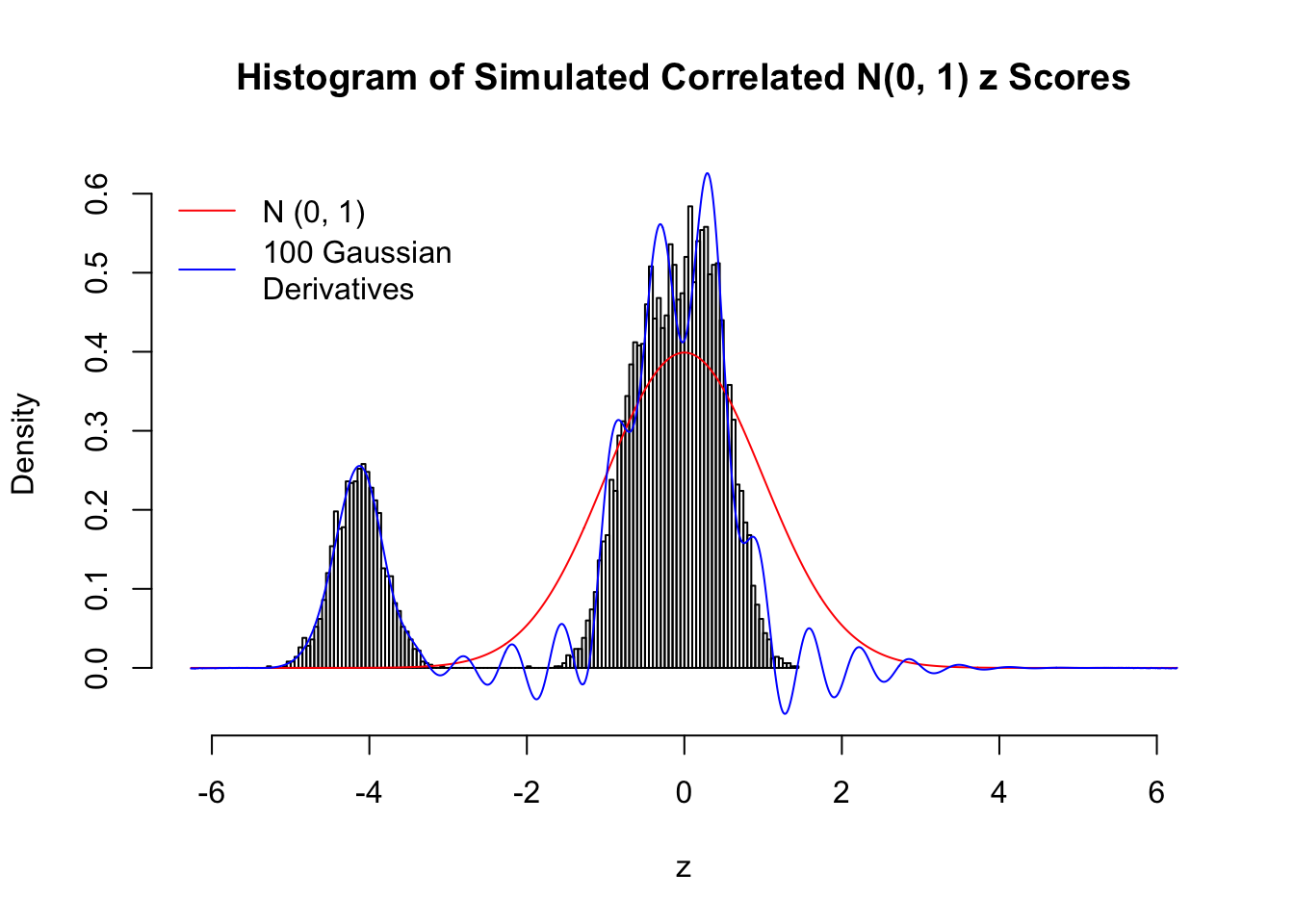
Despite his theory on the connection between Gaussian derivatives and empirical distributions of correlated null \(z\) scores, Dr. Schwartzman in his own research used Gaussian mixtures instead of Gaussian derivatives to fit the empirical distribution. A motivating example of his is a large number of marginally \(N\left(0, 1\right)\) \(z\) scores that are closely correlated with each other within one group, but independent between groups. We now show that data simulated in this way can also be fitted by Gaussian derivatives by the method of moments. To be specific, let \(n\) standard normal random samples be in \(K\) groups, in each group \(k\), given \(x_k\), \(y_{ki}\) iid \(N\left(0, 1\right)\),
\[ z_{ki} = \sqrt{\rho} x_k + \sqrt{1 - \rho} y_{ki} \ . \] In all the simulations, we choose \(n = 10^4\), \(\rho = 0.9\), and for theoretical exploration, \(L = 100\) Gaussian derivatives.
n = 1e4
rho = 0.9
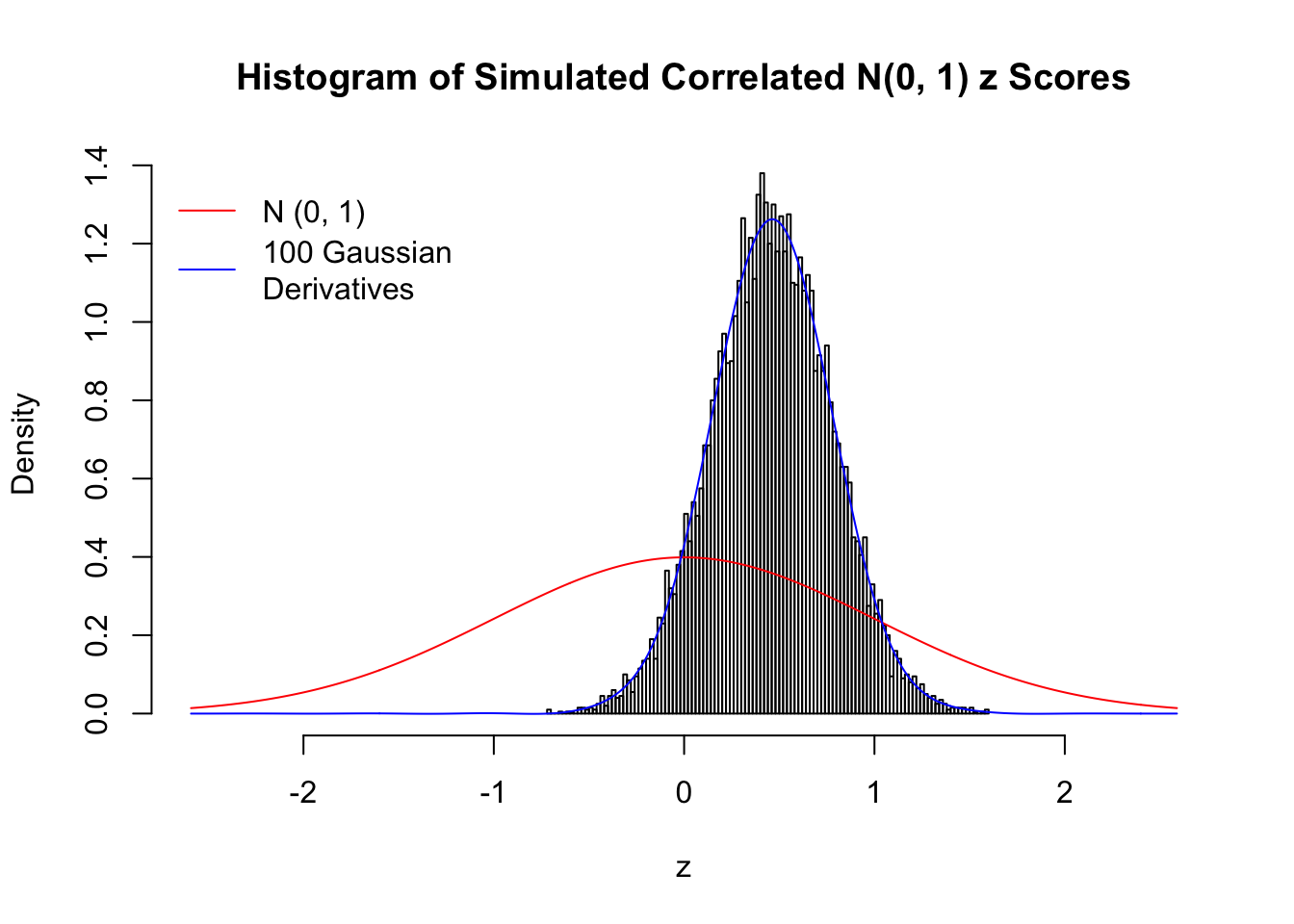
L = 100Scenario I: One Group
set.seed(777)
K = 1
for (j in 1 : 5) {
z = z.sim(n, K, rho)
fit.gd(L, z)
}
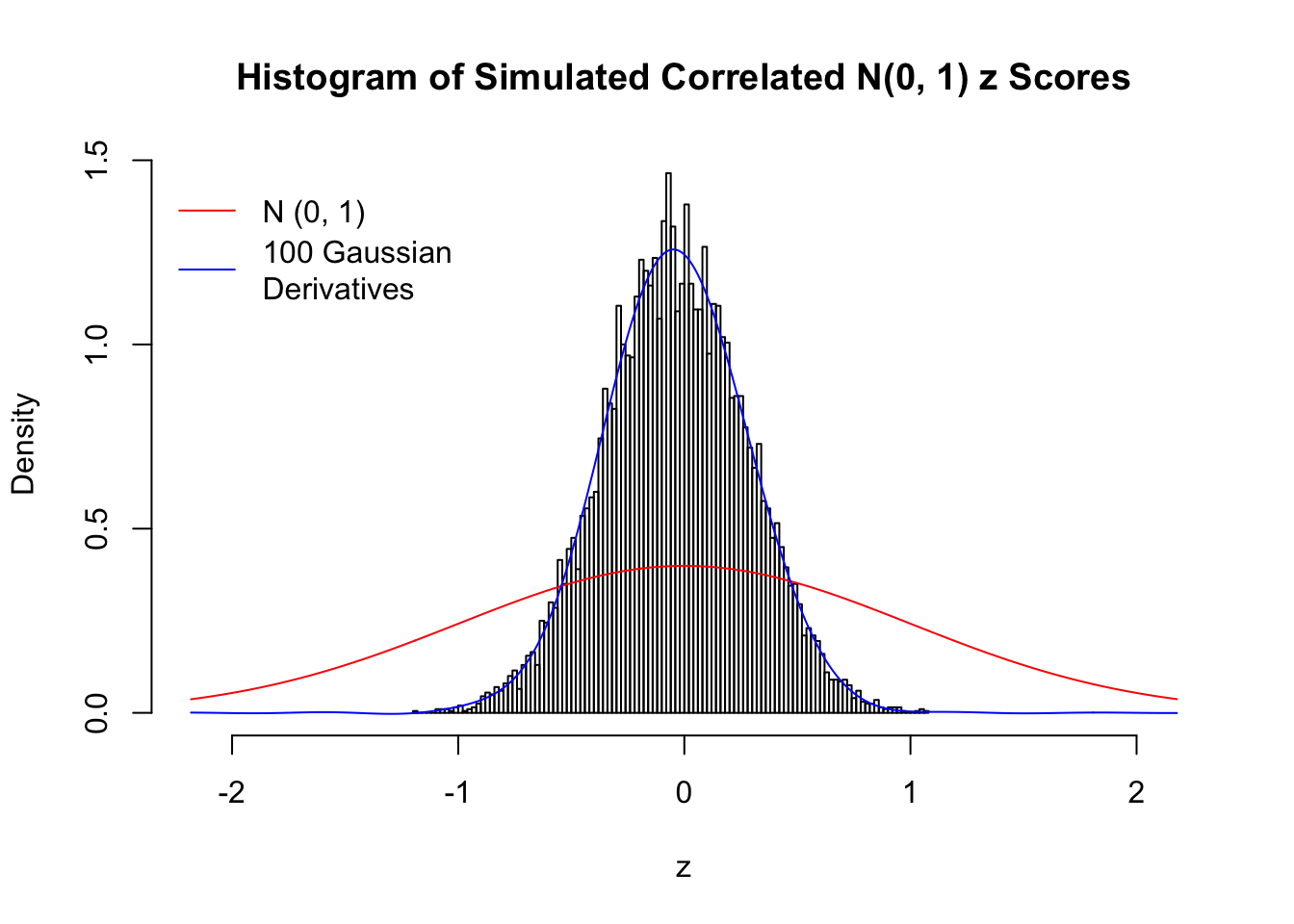
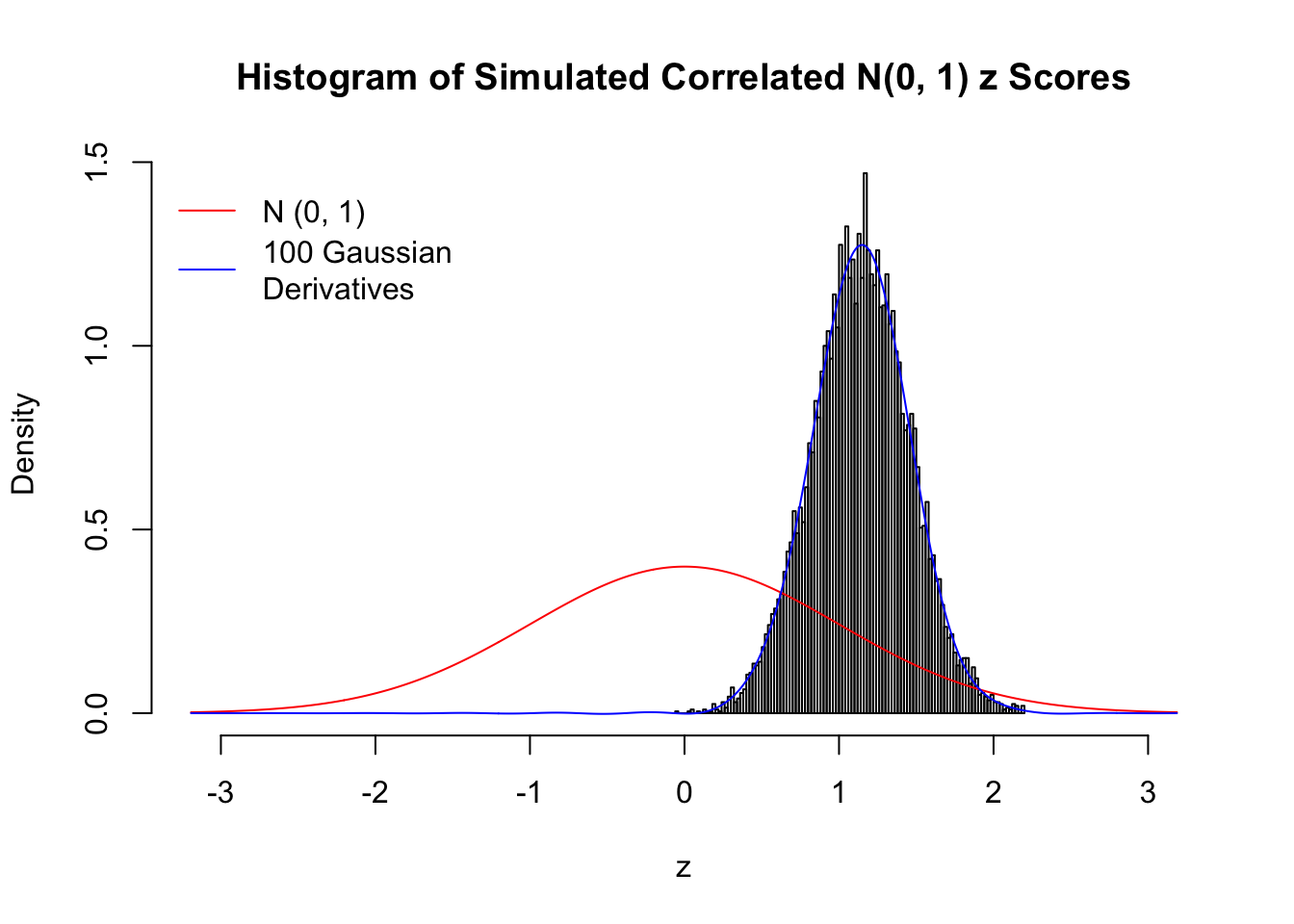
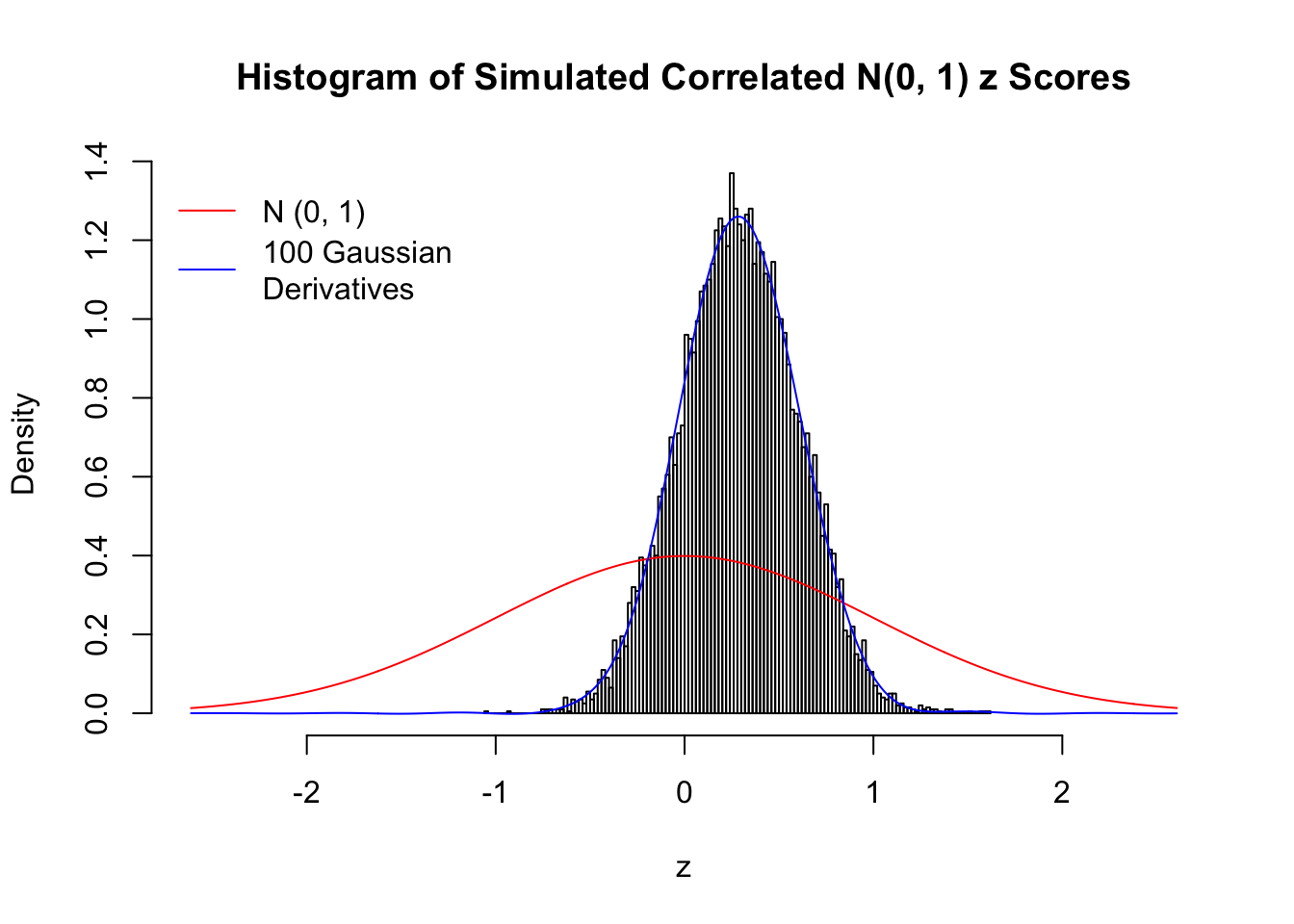
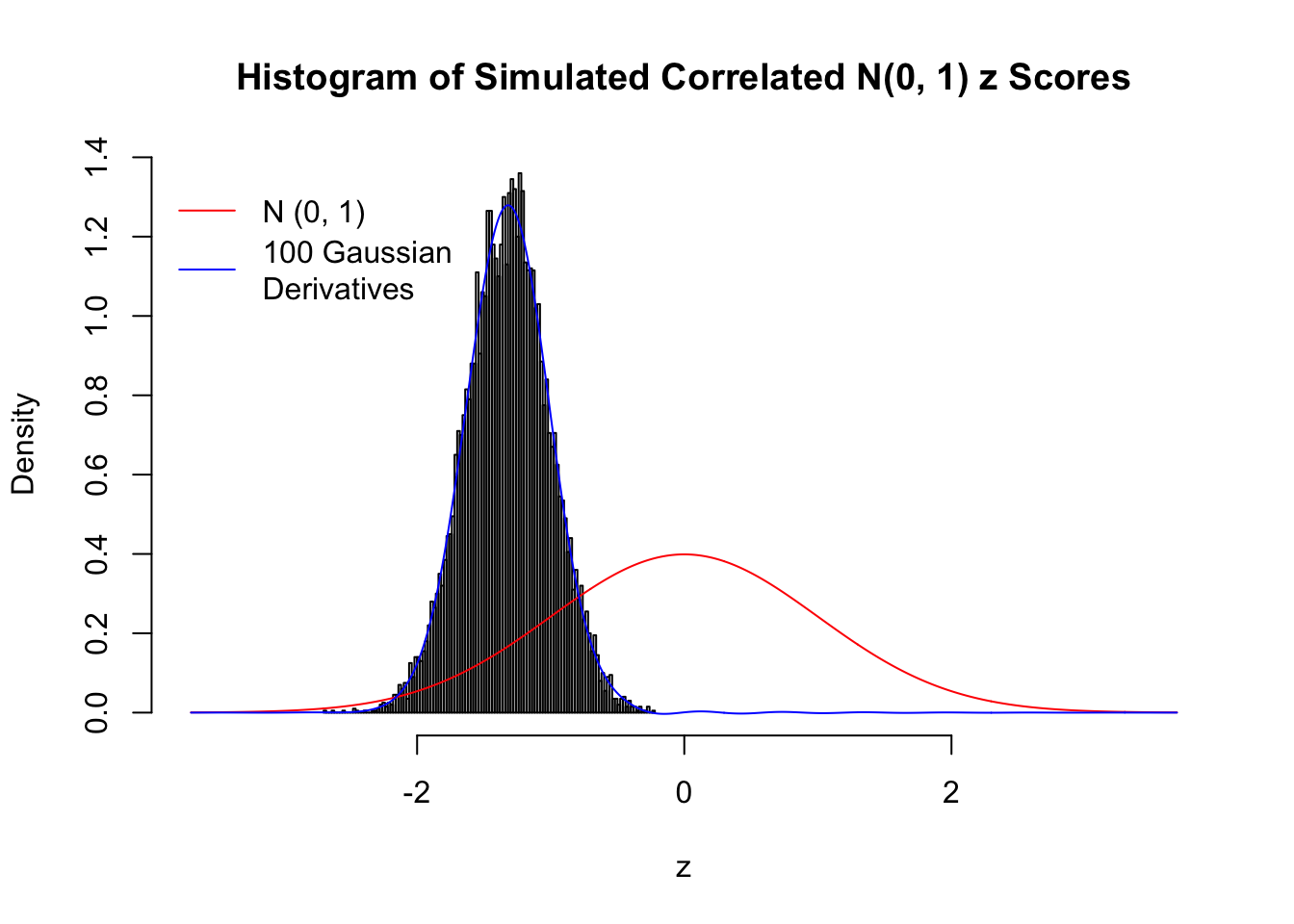
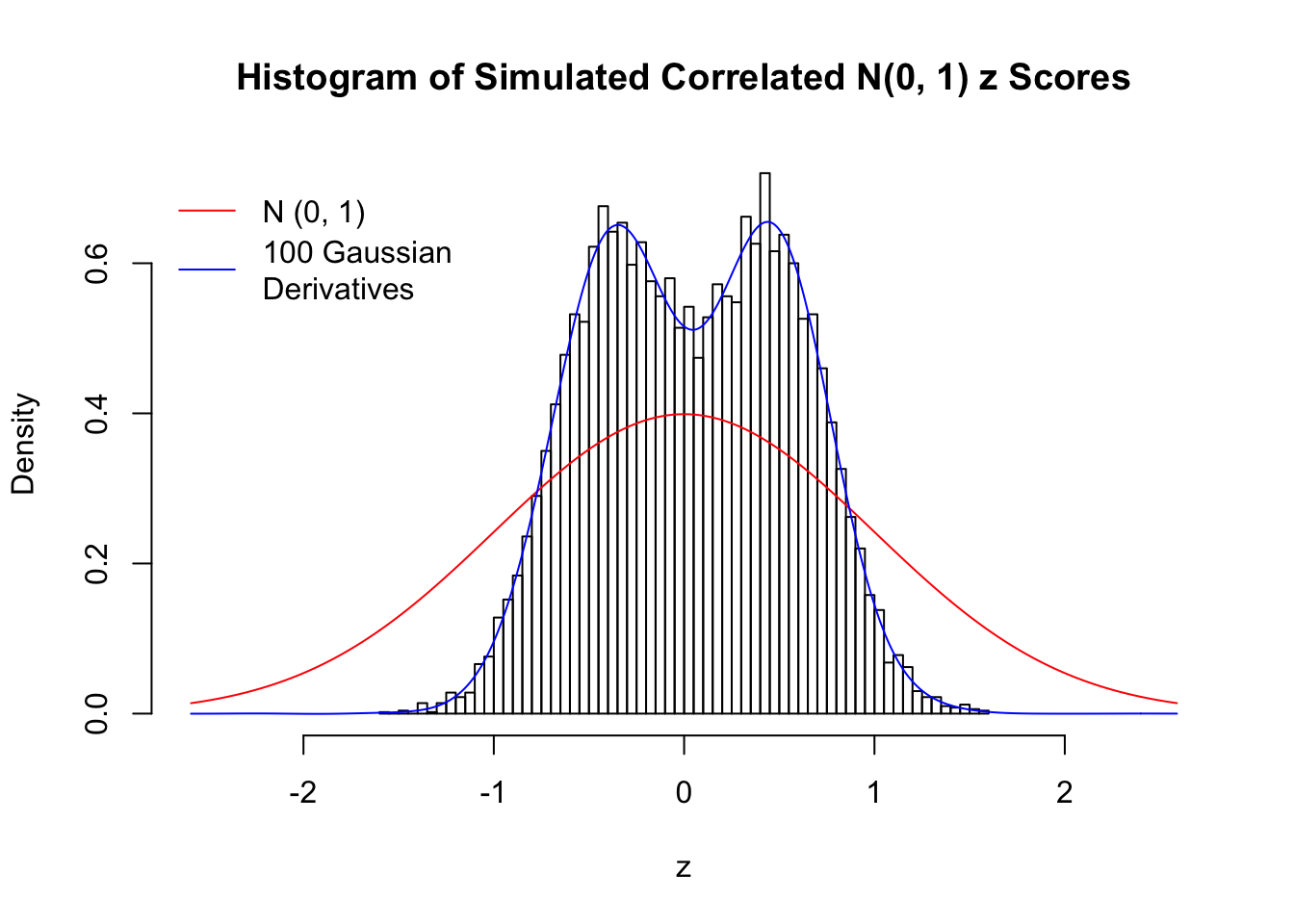
Scenario II: Two Groups
set.seed(777)
K = 2
for (j in 1 : 5) {
z = z.sim(n, K, rho)
fit.gd(L, z)
}
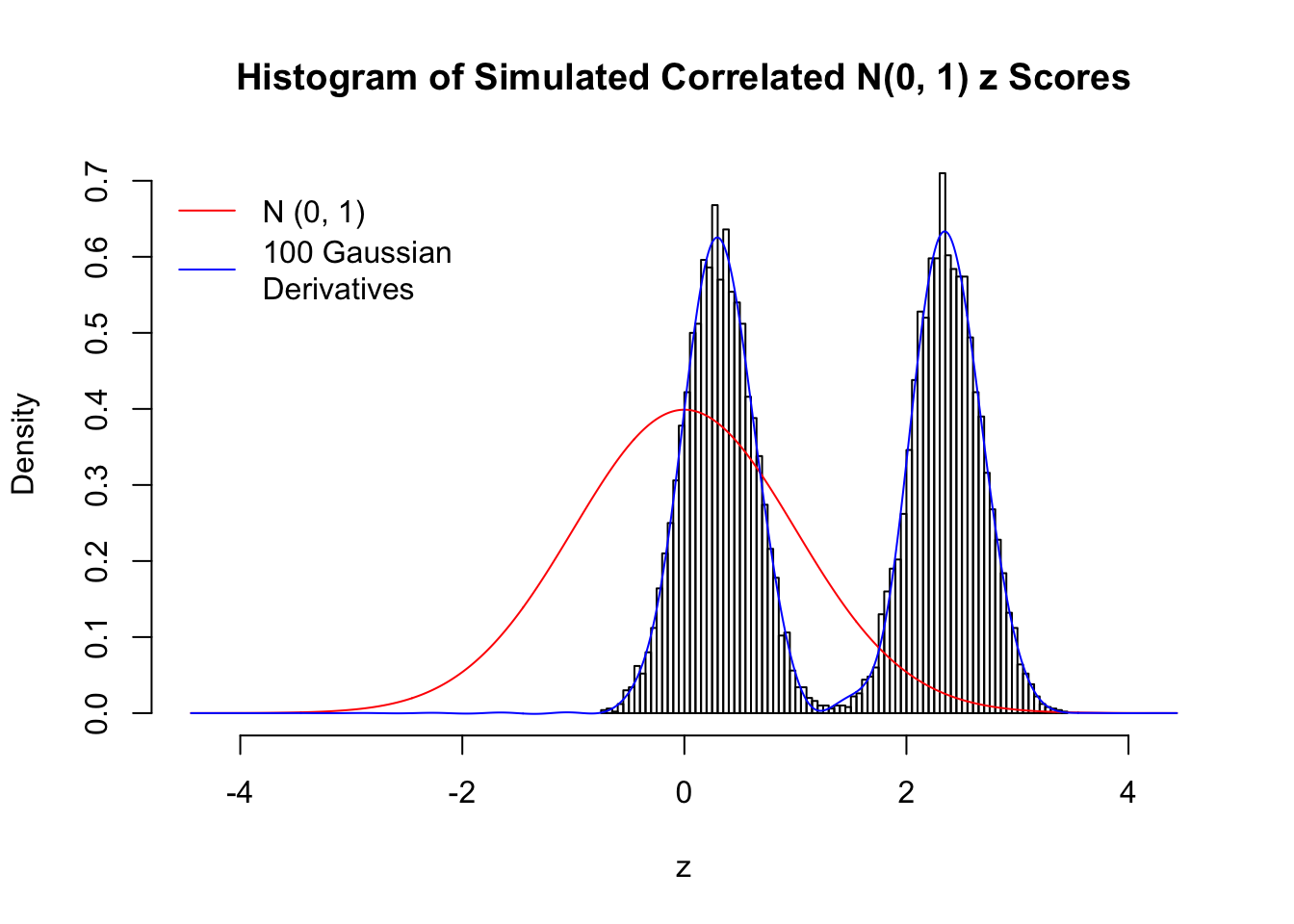
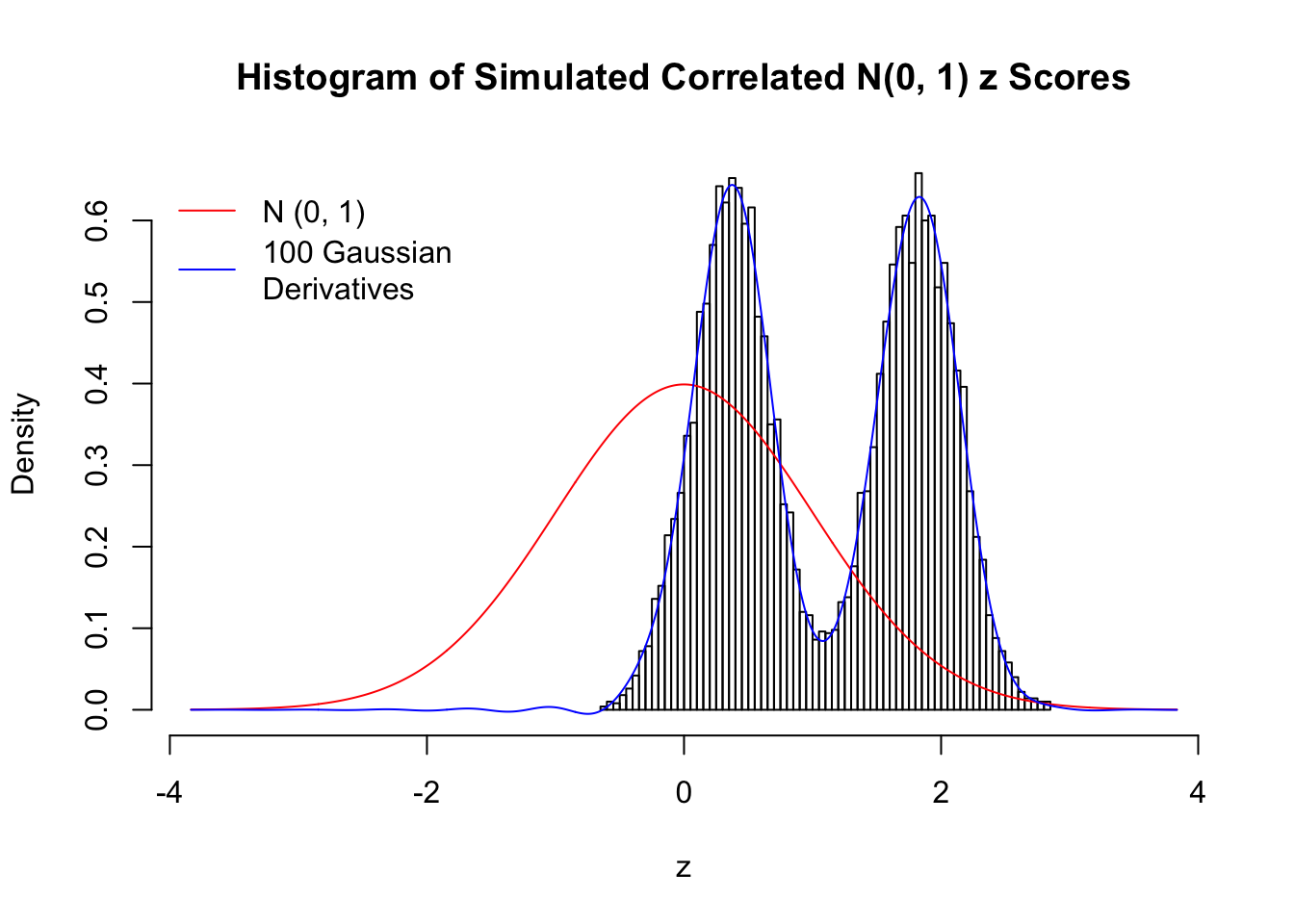
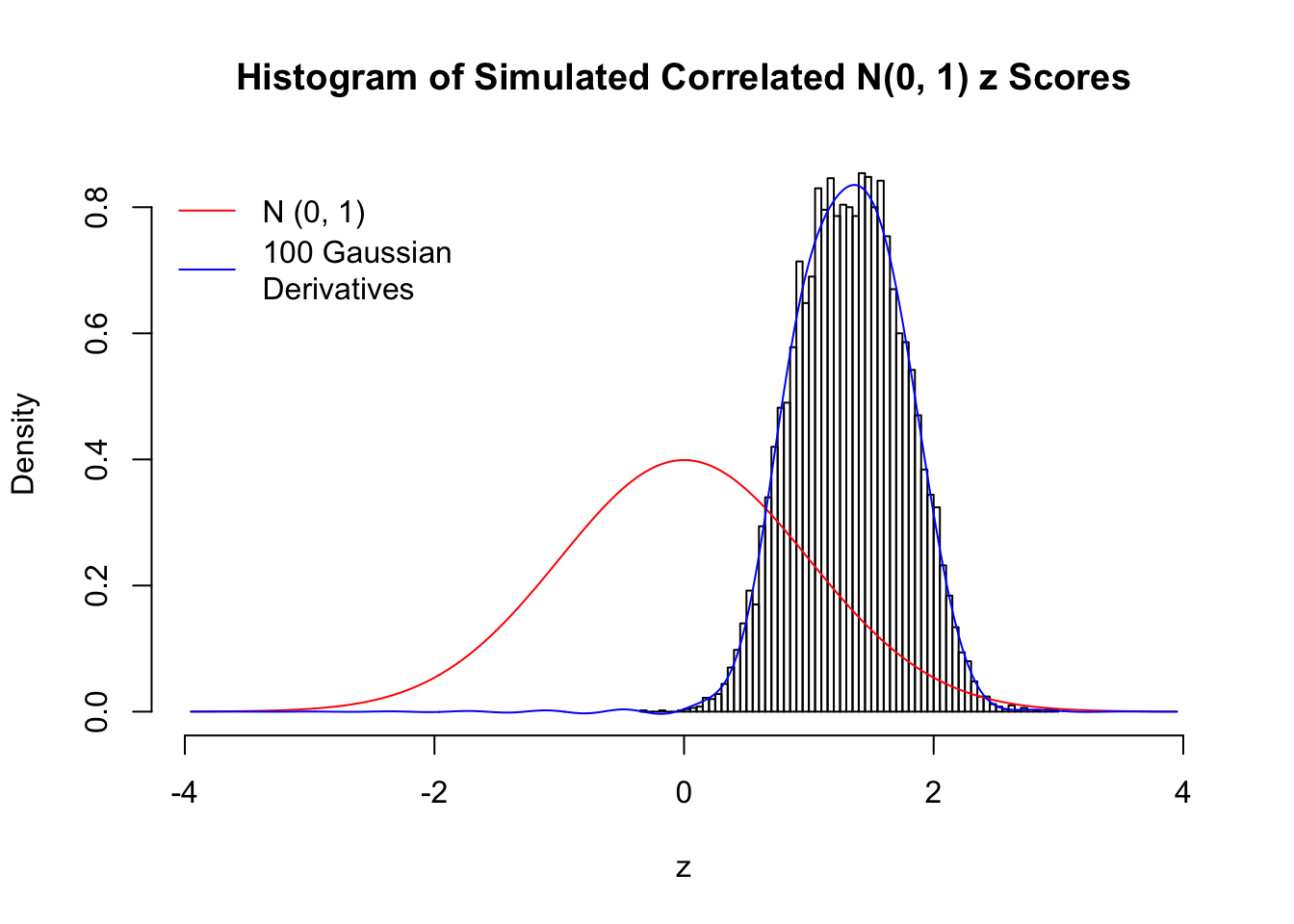
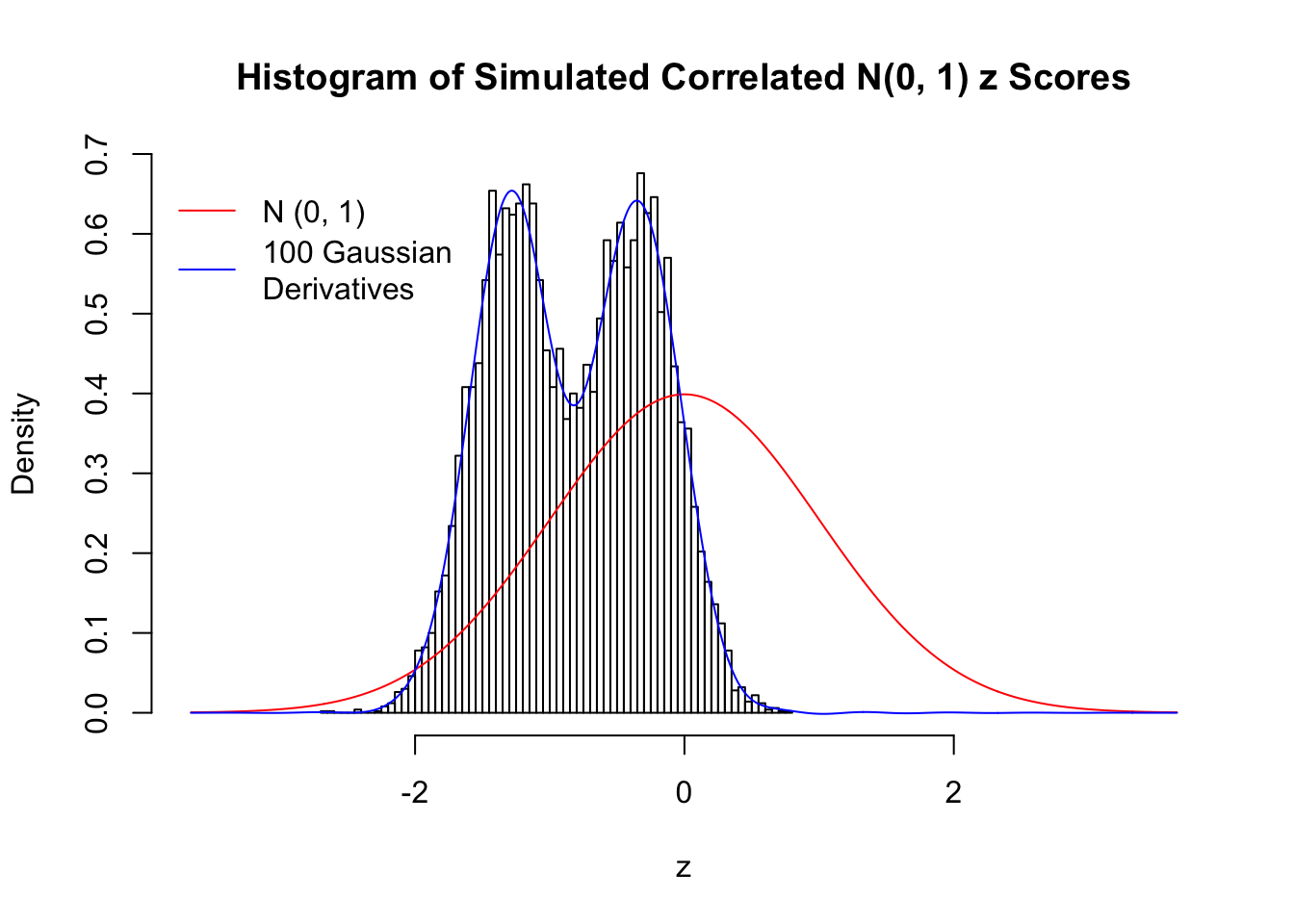
Scenario III: Three Groups
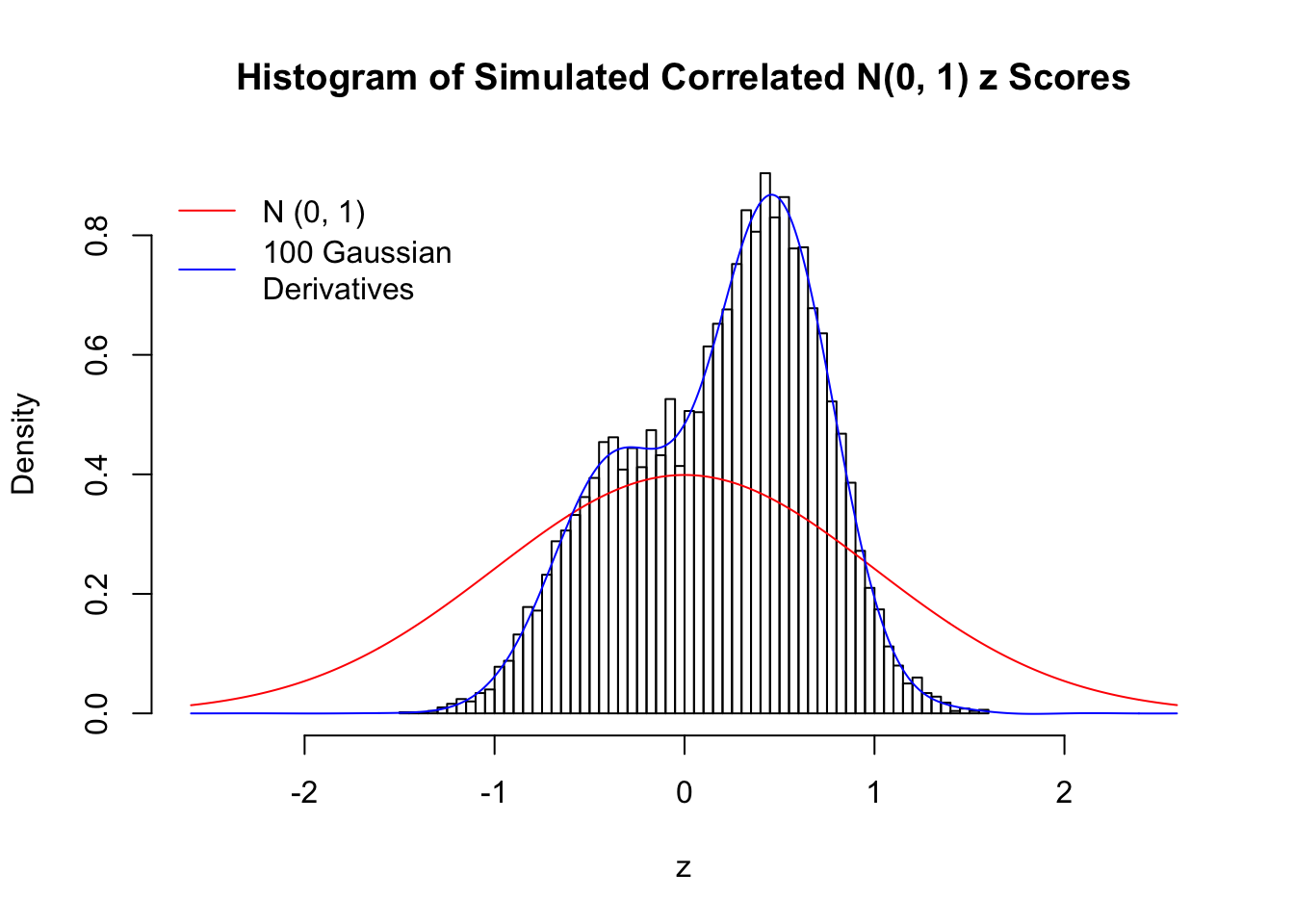
set.seed(777)
K = 3
for (j in 1 : 5) {
z = z.sim(n, K, rho)
fit.gd(L, z)
}
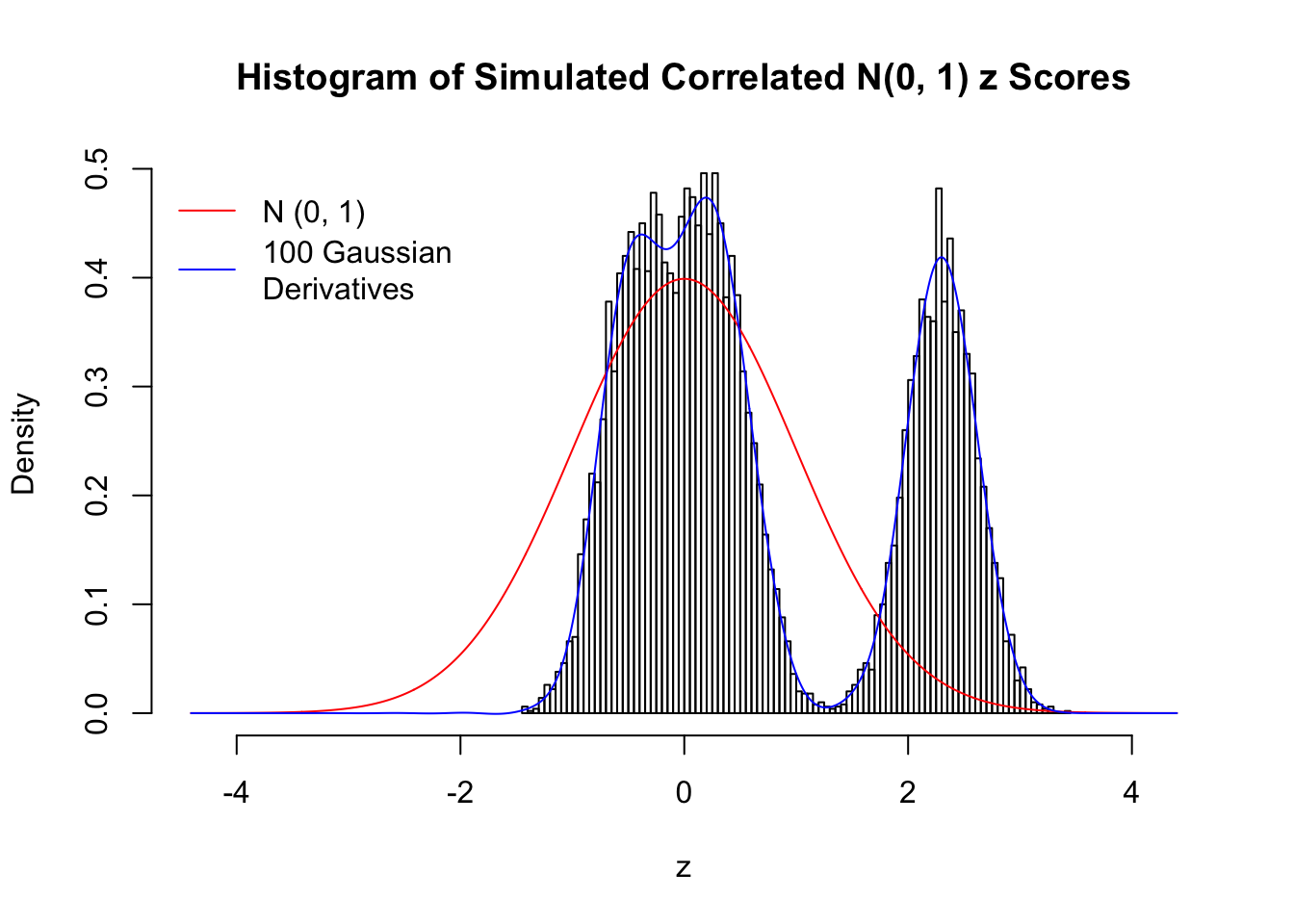
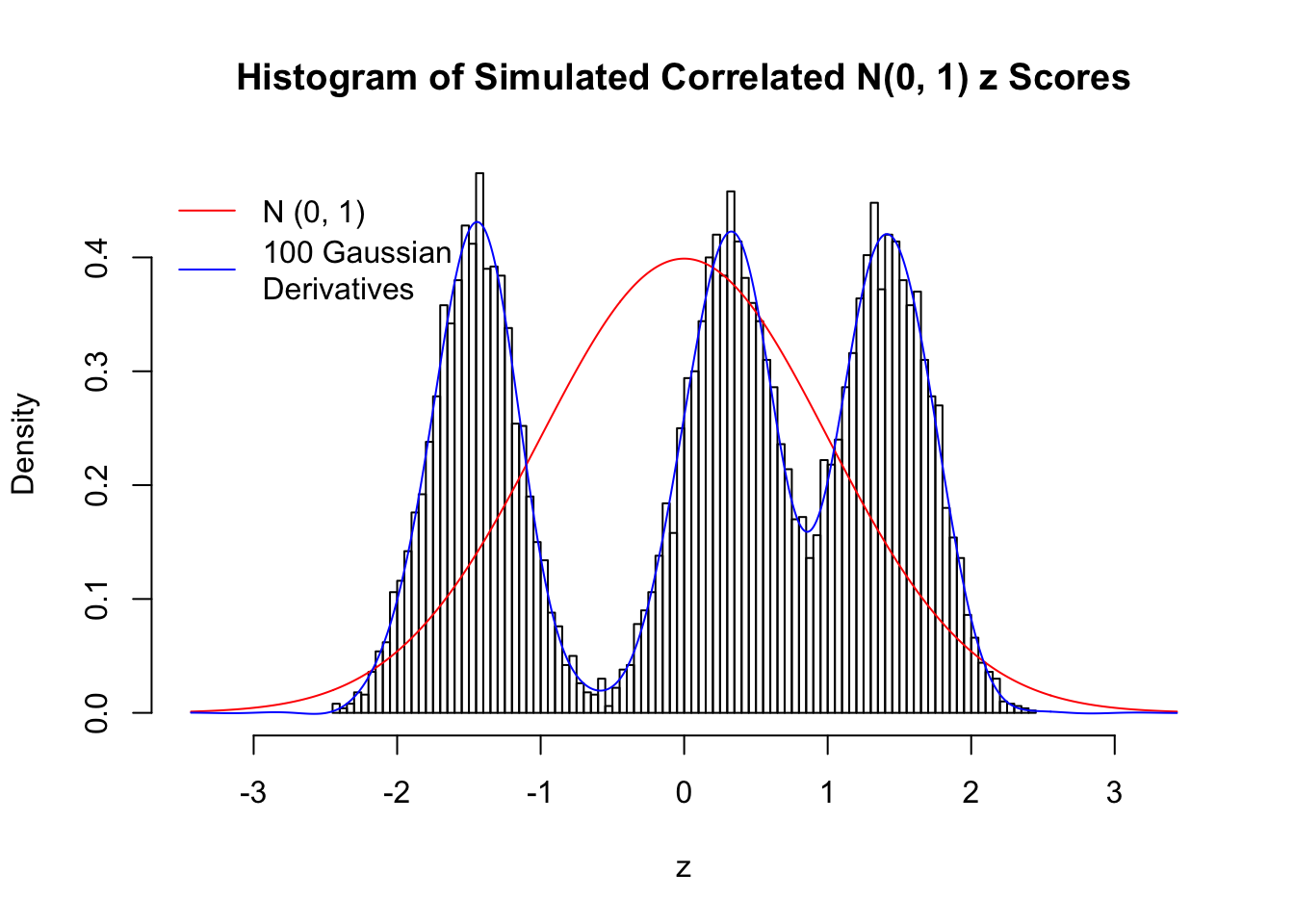
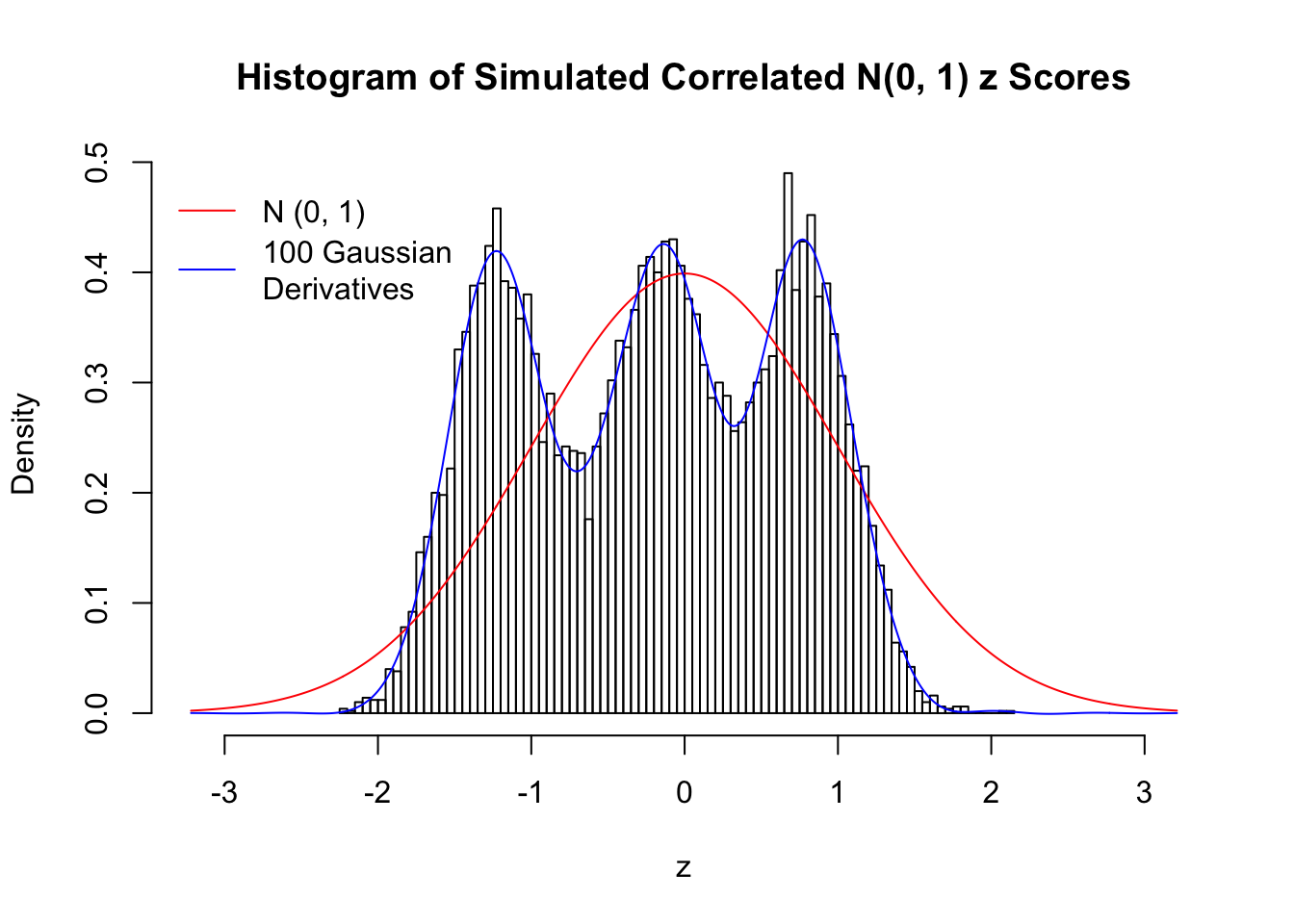
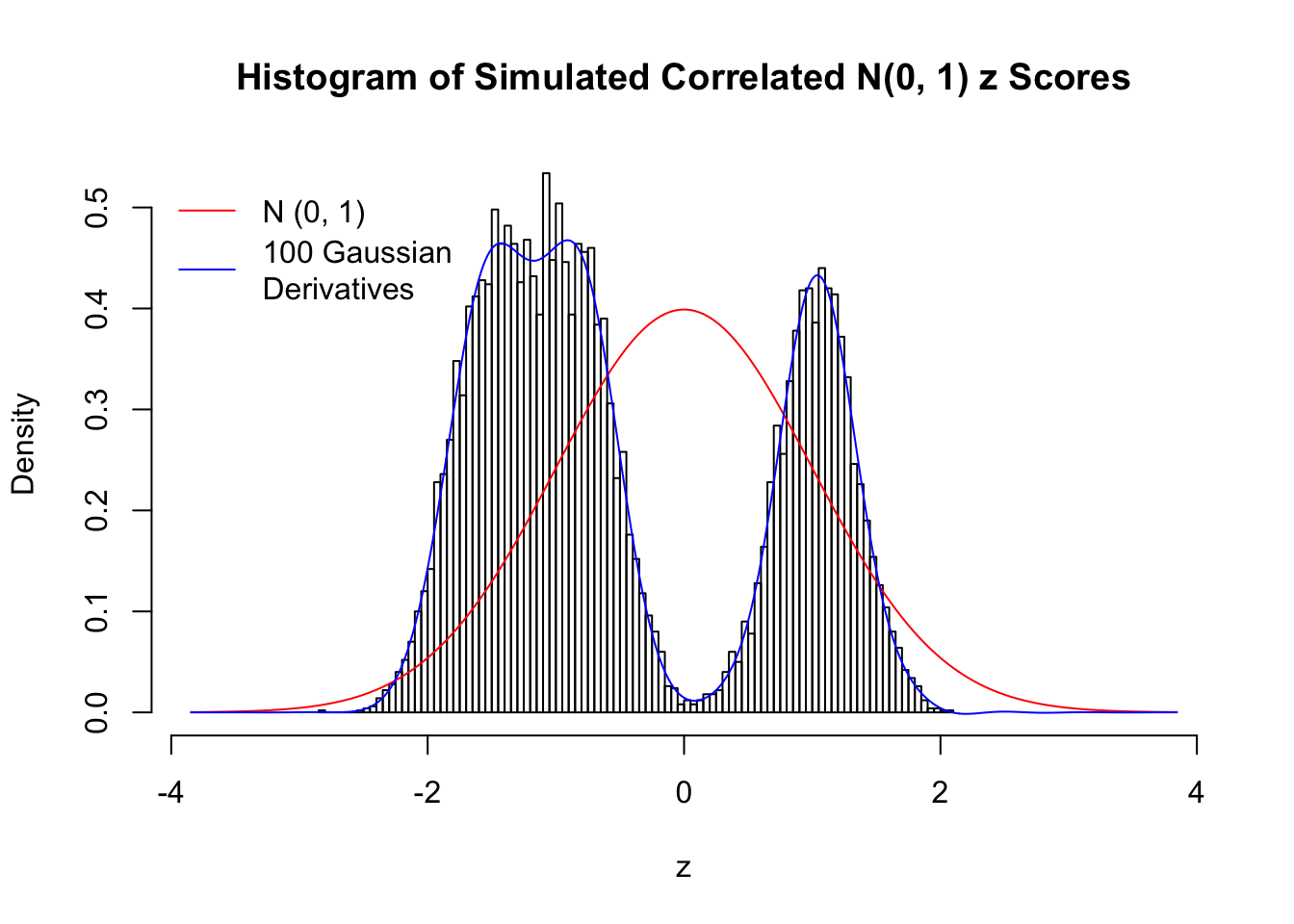
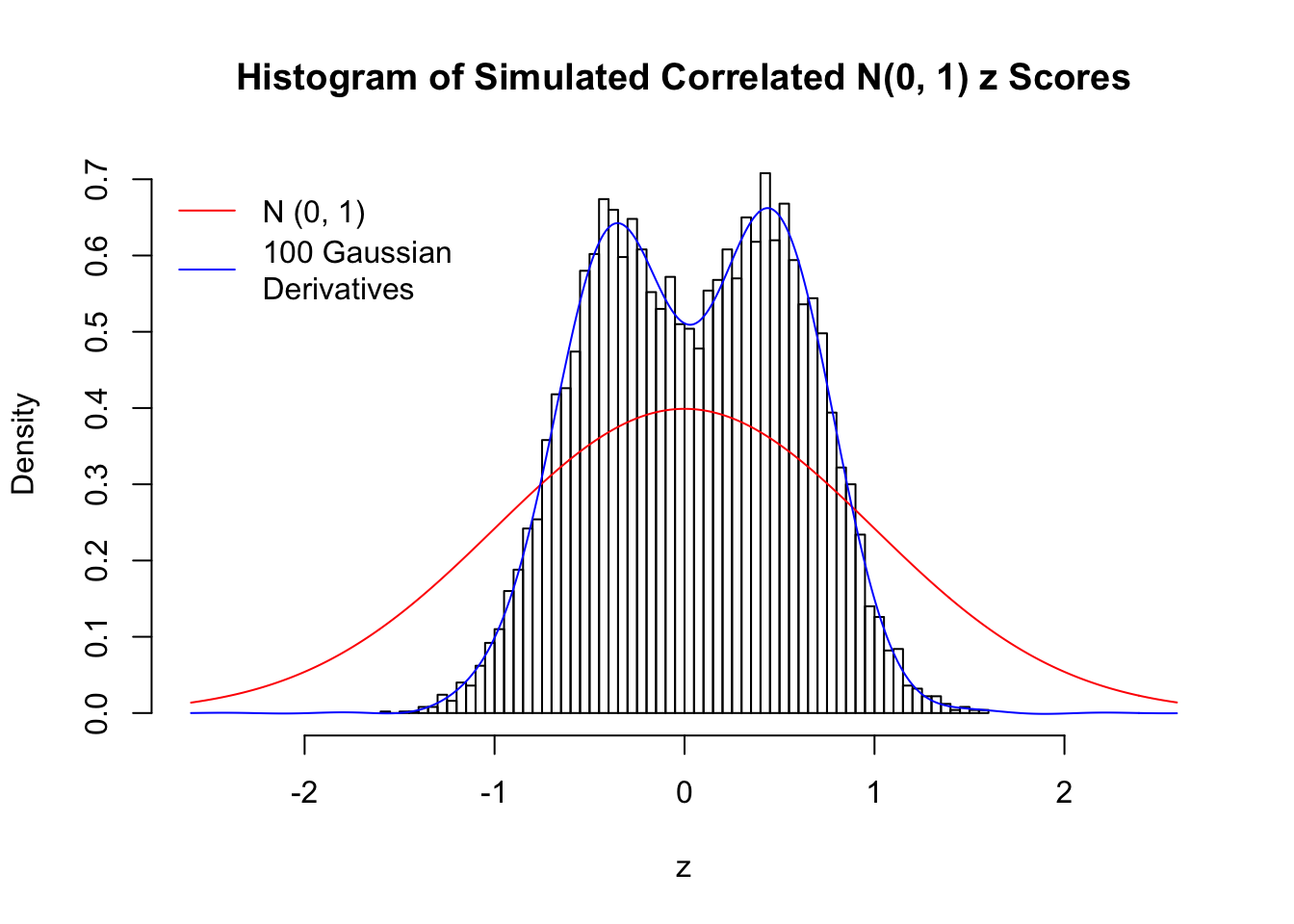
Scenario IV: Four Groups
set.seed(777)
K = 4
for (j in 1 : 5) {
z = z.sim(n, K, rho)
fit.gd(L, z)
}
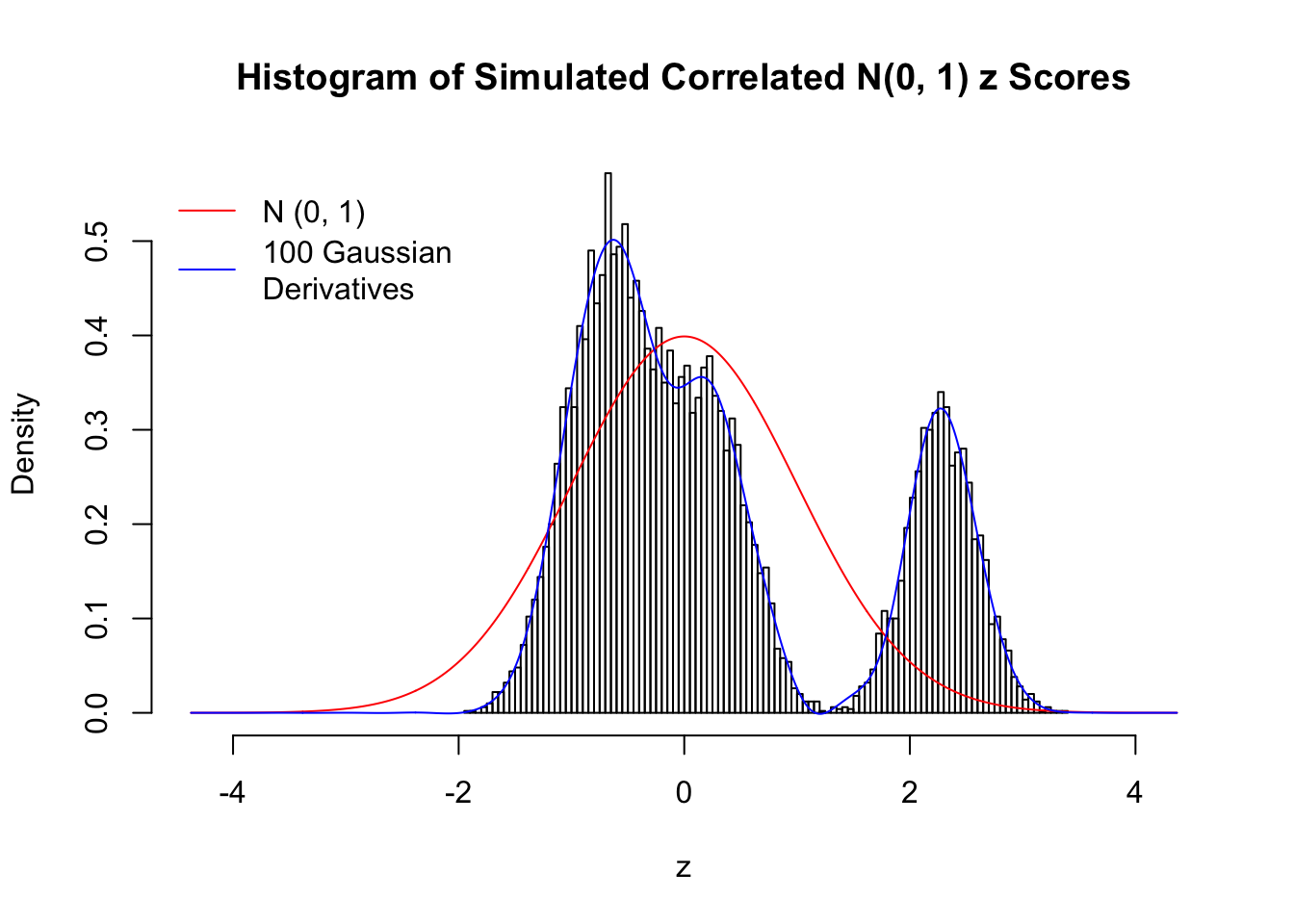
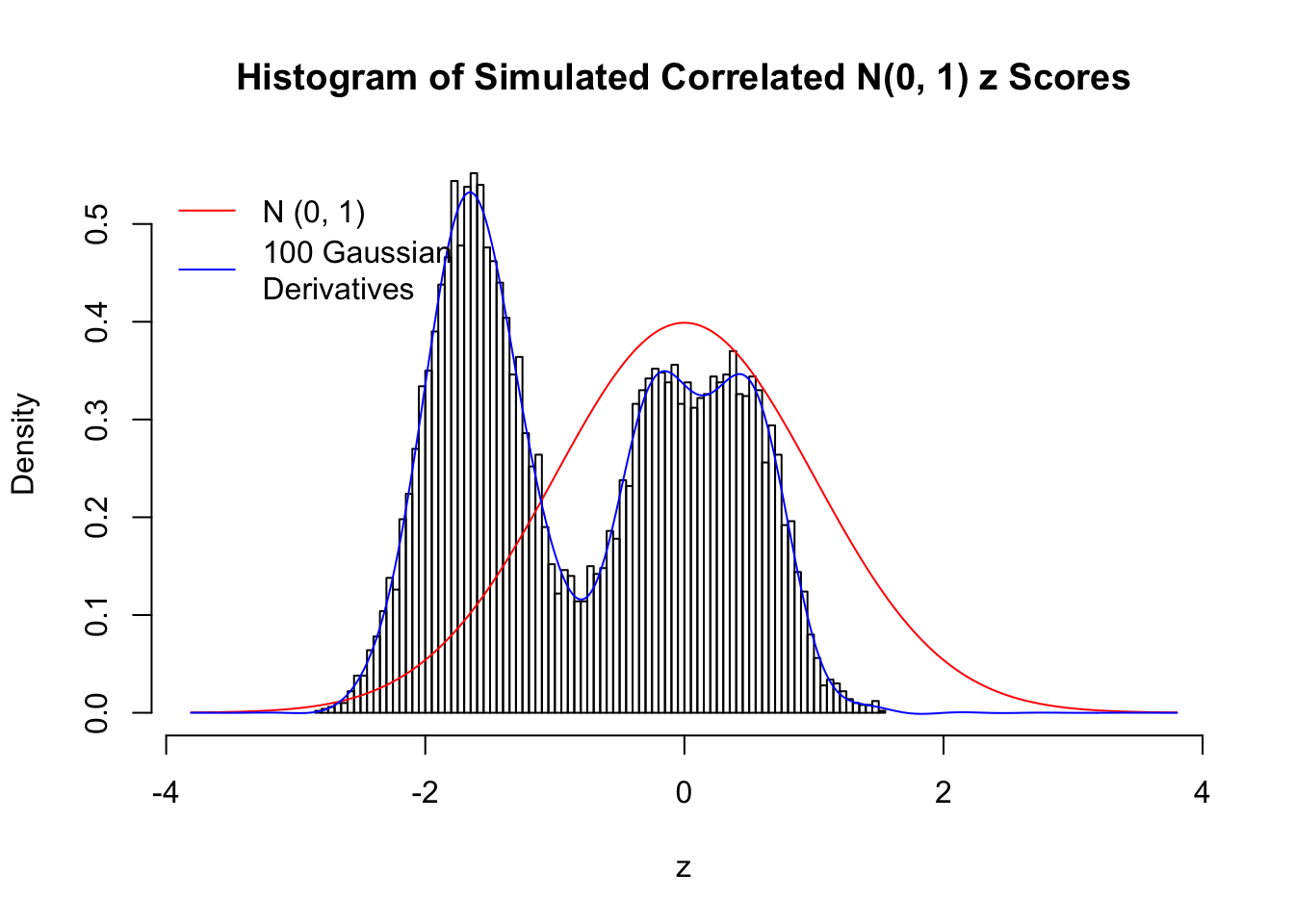
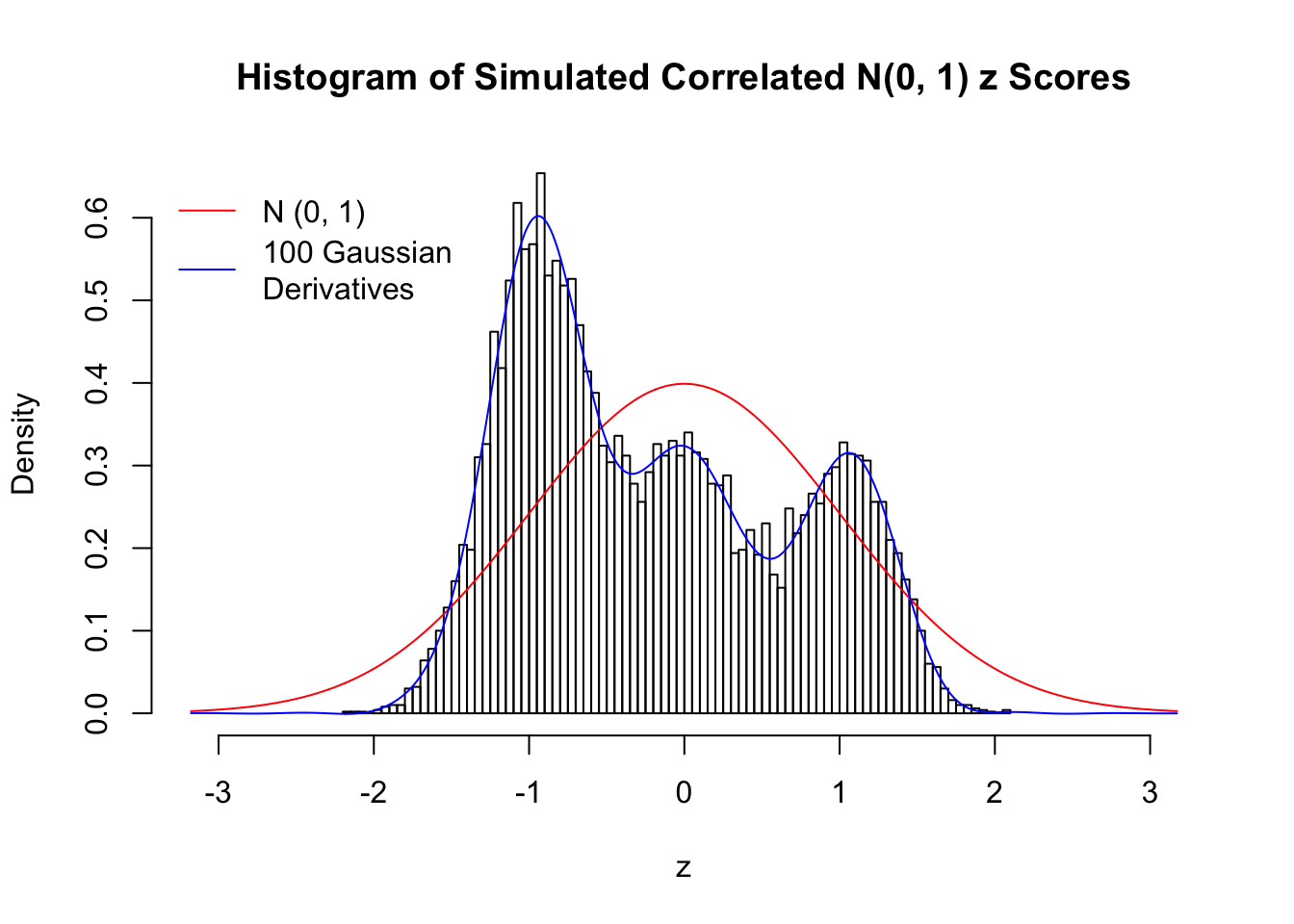
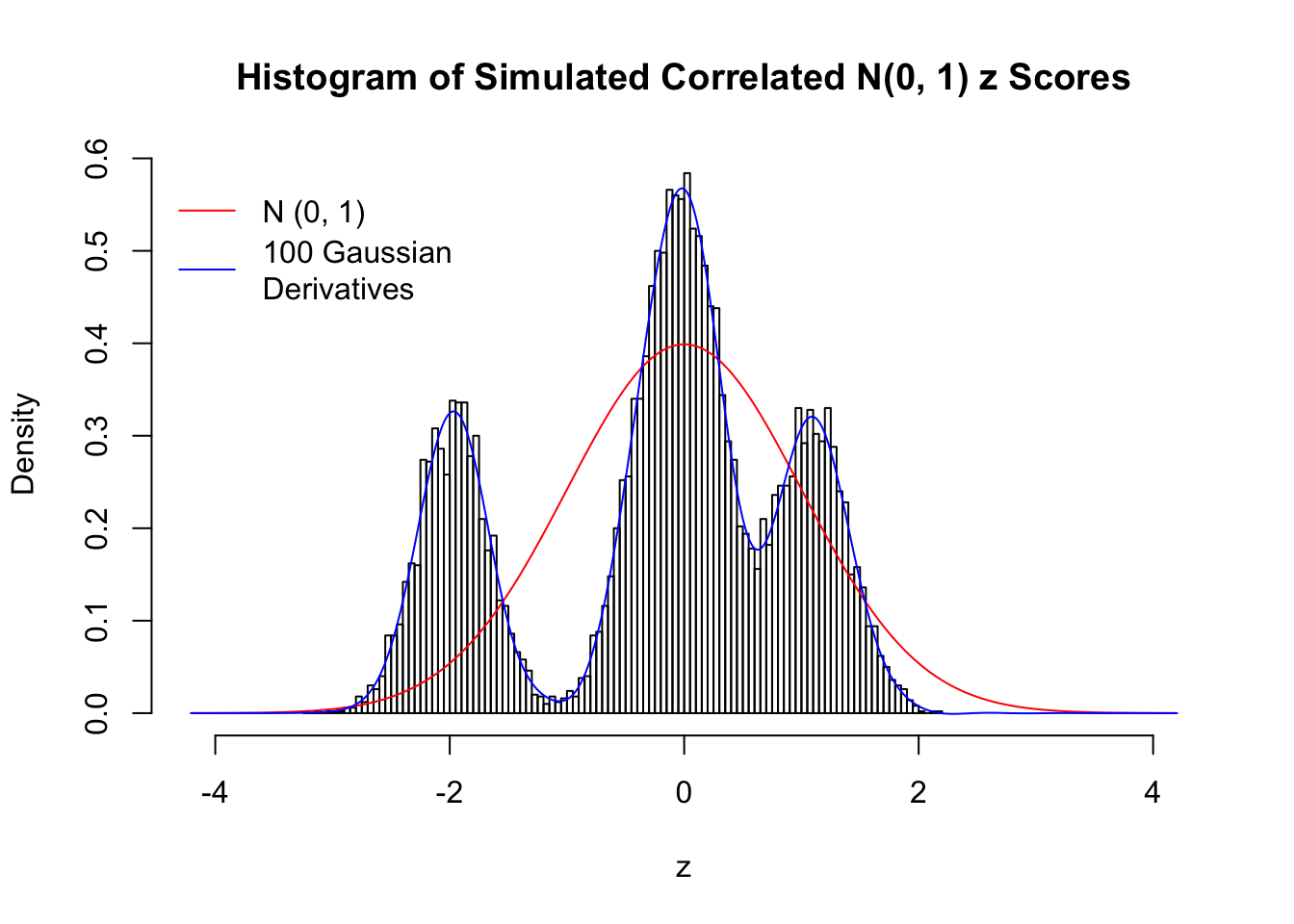
Scenario V: Five Groups
set.seed(777)
K = 5
for (j in 1 : 5) {
z = z.sim(n, K, rho)
fit.gd(L, z)
}
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_3.4.3 backports_1.1.2 magrittr_1.5 rprojroot_1.3-1
[5] tools_3.4.3 htmltools_0.3.6 yaml_2.1.16 Rcpp_0.12.14
[9] stringi_1.1.6 rmarkdown_1.8 knitr_1.17 git2r_0.20.0
[13] stringr_1.2.0 digest_0.6.13 evaluate_0.10.1This R Markdown site was created with workflowr